8FJ7
 
 | | LSD1-CoREST in complex with T108 and SNAG peptide | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, Zinc finger protein SNAI1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-12-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
8FRV
 
 | | LSD1-CoREST in complex with T17, short soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,13R)-3-[3-(dimethylcarbamoyl)phenyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-01-09 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To be published
|
|
8FRI
 
 | | LSD1-CoREST in complex with AW4, short soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3S,3aS,13R)-1-hydroxy-10,11-dimethyl-4,6-dioxo-3-([1~1~,2~1~:2~3~,3~1~-terphenyl]-1~4~-yl)-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-01-07 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8FRQ
 
 | | LSD1-CoREST in complex with T14, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(4aS)-7,8-dimethyl-5-(3-{4-[(5-methyl-1,3,4-thiadiazol-2-yl)carbamoyl]phenyl}propanoyl)-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-01-08 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
8FSK
 
 | | LSD1-CoREST in complex with T18, short soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,13R)-3-(3-benzamidophenyl)-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-01-10 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
2CKL
 
 | | Ring1b-Bmi1 E3 catalytic domain structure | | Descriptor: | IODIDE ION, POLYCOMB GROUP RING FINGER PROTEIN 4, UBIQUITIN LIGASE PROTEIN RING2, ... | | Authors: | Buchwald, G, van der Stoop, P, Weichenrieder, O, Perrakis, A, van Lohuizen, M, Sixma, T.K. | | Deposit date: | 2006-04-20 | | Release date: | 2006-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and E3-Ligase Activity of the Ring-Ring Complex of Polycomb Proteins Bmi1 and Ring1B.
Embo J., 25, 2006
|
|
3BRD
 
 | | CSL (Lag-1) bound to DNA with Lin-12 RAM peptide, P212121 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
4LCJ
 
 | | CtBP2 in complex with substrate MTOB | | Descriptor: | 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, C-terminal-binding protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hilbert, B.J, Schiffer, C.A, Royer Jr, W.E. | | Deposit date: | 2013-06-21 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structures of human CtBP in complex with substrate MTOB reveal active site features useful for inhibitor design.
Febs Lett., 588, 2014
|
|
3HC6
 
 | | FXR with SRC1 and GSK088 | | Descriptor: | 3-[(5-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-1H-indol-1-yl)methyl]benzoic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | FXR agonist activity of conformationally constrained analogs of GW 4064.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8TX8
 
 | |
6W1E
 
 | |
6W1F
 
 | |
8X19
 
 | | Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X15
 
 | | Structure of nucleosome-bound SRCAP-C in the apo state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X1C
 
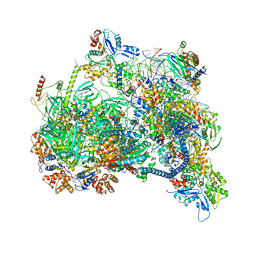 | | Structure of nucleosome-bound SRCAP-C in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | Deposit date: | 2023-11-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
3HC5
 
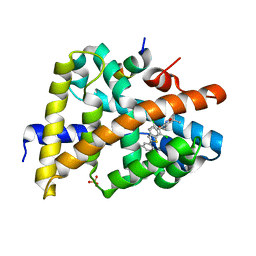 | | FXR with SRC1 and GSK826 | | Descriptor: | 3-(6-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-1-benzothiophen-2-yl)benzoic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | FXR agonist activity of conformationally constrained analogs of GW 4064.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4O8B
 
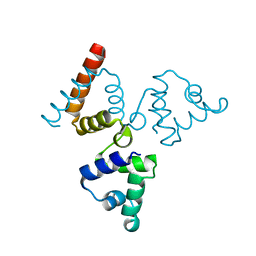 | | Crystal structure of transcriptional regulator BswR | | Descriptor: | Uncharacterized protein | | Authors: | Ye, F.Z, Wang, C, Kumar, V, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2013-12-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | BswR controls bacterial motility and biofilm formation in Pseudomonas aeruginosa through modulation of the small RNA rsmZ.
Nucleic Acids Res., 42, 2014
|
|
5YJB
 
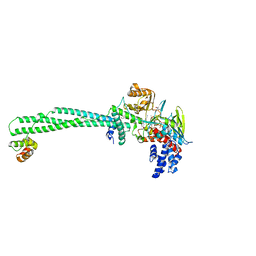 | | LSD1-CoREST in complex with 4-[5-(piperidin-4-ylmethoxy)-2-(p-tolyl)pyridin-3-yl]benzonitrile | | Descriptor: | 4-[2-(4-methylphenyl)-5-(piperidin-4-ylmethoxy)pyridin-3-yl]benzenecarbonitrile, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Hashimoto, T, Matsuno, K, Umehara, T. | | Deposit date: | 2017-10-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal Structure of LSD1 in Complex with 4-[5-(Piperidin-4-ylmethoxy)-2-(p-tolyl)pyridin-3-yl]benzonitrile.
Molecules, 23, 2018
|
|
4X0G
 
 | | Structure of Bsg25A binding with DNA | | Descriptor: | ACETATE ION, Blastoderm-specific gene 25A, DNA (5'-D(*GP*TP*TP*CP*CP*AP*AP*TP*TP*GP*GP*AP*A)-3') | | Authors: | Ren, A. | | Deposit date: | 2014-11-21 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2065 Å) | | Cite: | Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family.
Genes Dev., 29, 2015
|
|
6Y17
 
 | |
8AS9
 
 | | Crystal structure of the talin-KANK1 complex | | Descriptor: | B-cell lymphoma 6 protein, GLYCEROL, KN-motif NCoR1 BBD fusion,Nuclear receptor corepressor 1, ... | | Authors: | Zacharchenko, T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis of the talin-KANK1 interaction that coordinates the actin and microtubule cytoskeletons at focal adhesions.
Open Biology, 13, 2023
|
|
8DSZ
 
 | | PPARg bound to partial agonist H3B-487 | | Descriptor: | (2R)-2-{5-[(5-{[(1R)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DSY
 
 | | PPARg bound to inverse agonist H3B-343 | | Descriptor: | Peroxisome proliferator-activated receptor gamma, {5-[(5-{[(4-tert-butylphenyl)methyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}acetic acid | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
3VVZ
 
 | | Crystal Strucuture of The Rhodamine 6G-Bound Form of RamR (Transcriptional Regurator of TetR Family) from Salmonella Typhimurium | | Descriptor: | Putative regulatory protein, RHODAMINE 6G, SULFATE ION | | Authors: | Sakurai, K, Nikaido, E, Nakashima, R, Yamasaki, S, Yamaguchi, A, Nishino, K. | | Deposit date: | 2012-07-30 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The crystal structure of multidrug-resistance regulator RamR with multiple drugs
Nat Commun, 4, 2013
|
|
3VVY
 
 | | Crystal Strucuture of The Ethidium-Bound Form of RamR (Transcriptional Regurator of TetR Family) from Salmonella Typhimurium | | Descriptor: | ETHIDIUM, Putative regulatory protein | | Authors: | Sakurai, K, Nikaido, E, Nakashima, R, Yamasaki, S, Yamaguchi, A, Nishino, K. | | Deposit date: | 2012-07-30 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The crystal structure of multidrug-resistance regulator RamR with multiple drugs
Nat Commun, 4, 2013
|
|
