1XJ2
 
 | | CO-bound structure of bjFixLH | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, Sensor protein fixL | | Authors: | Key, J, Moffat, K. | | Deposit date: | 2004-09-22 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Deoxy and CO-Bound bjFixLH Reveal Details of Ligand Recognition and Signaling
Biochemistry, 44, 2005
|
|
5M3J
 
 | | Influenza B polymerase bound to four heptad repeats of serine 5 phosphorylated Pol II CTD | | Descriptor: | DNA-directed RNA polymerase subunit, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Lukarska, M, Pflug, A, Cusack, S. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of an essential interaction between influenza polymerase and Pol II CTD.
Nature, 541, 2017
|
|
4IQR
 
 | | Multi-Domain Organization of the HNF4alpha Nuclear Receptor Complex on DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), Hepatocyte nuclear factor 4-alpha, ... | | Authors: | Chandra, V, Huang, P, Kim, Y, Rastinejad, F. | | Deposit date: | 2013-01-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Multidomain integration in the structure of the HNF-4 alpha nuclear receptor complex.
Nature, 495, 2013
|
|
3LQF
 
 | | Crystal structure of the short-chain dehydrogenase Galactitol-Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD and erythritol | | Descriptor: | Galactitol dehydrogenase, MAGNESIUM ION, MESO-ERYTHRITOL, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Zander, U, Klink, B.U, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into substrate differentiation of the sugar-metabolizing enzyme galactitol dehydrogenase from Rhodobacter sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
3OX1
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-{2-[7-(methylsulfamoyl)naphthalen-1-yl]ethyl}acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
4IRR
 
 | | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP
To be Published
|
|
3LWC
 
 | |
4ISK
 
 | | Crystal structure of E.coli thymidylate synthase with dUMP and the BGC 945 inhibitor | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2'-deoxy-5'-uridylic acid, MAGNESIUM ION, ... | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2013-01-16 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Development and Binding Mode Assessment of N-[4-[2-Propyn-1-yl[(6S)-4,6,7,8-tetrahydro-2-(hydroxymethyl)-4-oxo-3H-cyclopenta[g]quinazolin-6-yl]amino]benzoyl]-l-gamma-glutamyl-d-glutamic Acid (BGC 945), a Novel Thymidylate Synthase Inhibitor That Targets Tumor Cells.
J.Med.Chem., 56, 2013
|
|
1XIS
 
 | |
4H1N
 
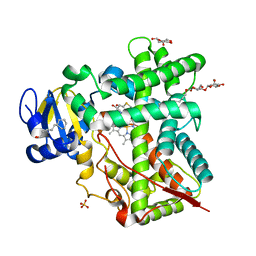 | | Crystal Structure of P450 2B4 F297A Mutant in Complex with Anti-platelet Drug Clopidogrel | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Clopidogrel, Cytochrome P450 2B4, ... | | Authors: | Shah, M.B, Jang, H.H, Stout, C.D, Halpert, J.R. | | Deposit date: | 2012-09-10 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | X-ray crystal structure of the cytochrome P450 2B4 active site mutant F297A in complex with clopidogrel: Insights into compensatory rearrangements of the binding pocket.
Arch.Biochem.Biophys., 530, 2013
|
|
5LVP
 
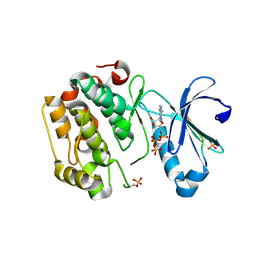 | | Human PDK1 Kinase Domain in Complex with an HM-Peptide Bound to the PIF-Pocket | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schulze, J.O, Saladino, G, Busschots, K, Neimanis, S, Suess, E, Odadzic, D, Zeuzem, S, Hindie, V, Herbrand, A.K, Lisa, M.N, Alzari, P.M, Gervasio, F.L, Biondi, R.M. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bidirectional Allosteric Communication between the ATP-Binding Site and the Regulatory PIF Pocket in PDK1 Protein Kinase.
Cell Chem Biol, 23, 2016
|
|
7BOG
 
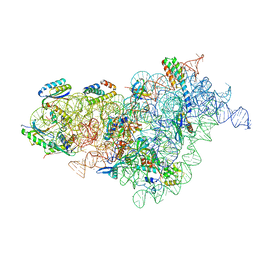 | | Bacterial 30S ribosomal subunit assembly complex state E (body domain) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
3P3Z
 
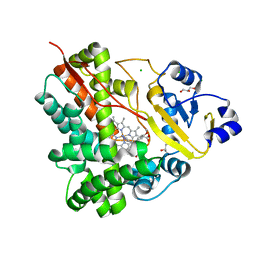 | | Crystal Structure of the Cytochrome P450 Monooxygenase AurH from Streptomyces Thioluteus in Complex with Ancymidol | | Descriptor: | (S)-cyclopropyl(4-methoxyphenyl)pyrimidin-5-ylmethanol, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zocher, G, Richter, M.E.A, Mueller, U, Hertweck, C. | | Deposit date: | 2010-10-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural fine-tuning of a multifunctional cytochrome p450 monooxygenase.
J.Am.Chem.Soc., 133, 2011
|
|
7BOD
 
 | | Bacterial 30S ribosomal subunit assembly complex state M (body domain) | | Descriptor: | 16S rRNA (body domain of 30S subunit), 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S.R. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
3I41
 
 | |
3I48
 
 | | Crystal structure of beta toxin from Staphylococcus aureus F277A, P278A mutant with bound magnesium ions | | Descriptor: | Beta-hemolysin, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Huseby, M, Shi, K, Kruse, A.C, Ohlendorf, D.H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biological functions of beta toxin from Staphylococcus aureus: Role of the hydrophobic beta hairpin in virulence
to be published, 2009
|
|
4S20
 
 | | Structural basis for transcription reactivation by RapA | | Descriptor: | 5'-D(P*AP*CP*GP*AP*CP*TP*GP*AP*GP*CP*CP*GP*AP*TP*G)-3', 5'-R(P*AP*UP*CP*GP*GP*CP*UP*CP*A)-3', DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Liu, B, Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-01-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Structural basis for transcription reactivation by RapA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1Y4C
 
 | | Designed Helical Protein fusion MBP | | Descriptor: | Maltose binding protein fused with designed helical protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | LaPorte, S.L, Forsyth, C.M, Cunningham, B.C, Miercke, L.J, Akhavan, D, Stroud, R.M. | | Deposit date: | 2004-11-30 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo design of an IL-4 antagonist and its structure at 1.9 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5PO6
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10157a | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-bromophenyl)-1H-pyrazol-3-amine, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3N2C
 
 | | Crystal structure of prolidase eah89906 complexed with n-methylphosphonate-l-proline | | Descriptor: | 1-[(R)-hydroxy(methyl)phosphoryl]-L-proline, PROLIDASE, ZINC ION | | Authors: | Patskovsky, Y, Xu, C, Sauder, J.M, Burley, S.K, Raushel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
5NZR
 
 | | The structure of the COPI coat leaf | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5O35
 
 | | Structure of complement proteins complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, Complement factor H,Complement factor H | | Authors: | Xue, X, Wu, J, Forneris, F, Gros, P. | | Deposit date: | 2017-05-23 | | Release date: | 2017-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7BKB
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (hexameric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
7BKC
 
 | | Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (dimeric, composite structure) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CoB--CoM heterodisulfide reductase iron-sulfur subunit A, CoB--CoM heterodisulfide reductase subunit B, ... | | Authors: | Pfeil-Gardiner, O, Watanabe, T, Shima, S, Murphy, B.J. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Three-megadalton complex of methanogenic electron-bifurcating and CO 2 -fixing enzymes.
Science, 373, 2021
|
|
5POO
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10966a | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-acetylpiperazin-1-yl)pyridine-3-carbonitrile, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
