4U3N
 
 | | Crystal structure of CCA trinucleotide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U55
 
 | | Crystal structure of Cryptopleurine bound to the yeast 80S ribosome | | Descriptor: | (14aR)-2,3,6-trimethoxy-11,12,13,14,14a,15-hexahydro-9H-dibenzo[f,h]pyrido[1,2-b]isoquinoline, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4UG0
 
 | | STRUCTURE OF THE HUMAN 80S RIBOSOME | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S RIBOSOMAL PROTEIN, ... | | Authors: | Khatter, H, Myasnikov, A.G, Natchiar, S.K, Klaholz, B.P. | | Deposit date: | 2015-03-20 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the human 80S ribosome
NATURE, 520, 2015
|
|
4U3U
 
 | | Crystal structure of Cycloheximide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4UJE
 
 | | Regulation of the mammalian elongation cycle by 40S subunit rolling: a eukaryotic-specific ribosome rearrangement | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S RIBOSOMAL PROTEIN S10, ... | | Authors: | Budkevich, T.V, Giesebrecht, J, Behrmann, E, Loerke, J, Ramrath, D.J.F, Mielke, T, Ismer, J, Hildebrand, P, Tung, C.-S, Nierhaus, K.H, Sanbonmatsu, K.Y, Spahn, C.M.T. | | Deposit date: | 2014-04-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Regulation of the Mammalian Elongation Cycle by Subunit Rolling: A Eukaryotic-Specific Ribosome Rearrangement.
Cell(Cambridge,Mass.), 158, 2014
|
|
4U6F
 
 | | Crystal structure of T-2 toxin bound to the yeast 80S ribosome | | Descriptor: | 12,13-Epoxytrichothec-9-ene-3,4,8,15-tetrol-4,15-diacetate-8-isovalerate, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-28 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U4U
 
 | | Crystal structure of Lycorine bound to the yeast 80S ribosome | | Descriptor: | (1S,2S,12bS,12cS)-2,4,5,7,12b,12c-hexahydro-1H-[1,3]dioxolo[4,5-j]pyrrolo[3,2,1-de]phenanthridine-1,2-diol, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U4Z
 
 | | Crystal structure of Phyllanthoside bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
8Q5I
 
 | | Structure of Candida albicans 80S ribosome in complex with cephaeline | | Descriptor: | 18S ribosomal RNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Atamas, A, Guskov, A, Yusupov, M. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural characterization of cephaeline binding to the eukaryotic ribosome using Cryo-Electron Microscopy
Biopolym Cell, 2023
|
|
4UJD
 
 | | mammalian 80S HCV-IRES initiation complex with eIF5B PRE-like state | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Eif5B on Hcv Ires
Nat.Struct.Mol.Biol., 21, 2014
|
|
8PPK
 
 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
8PPL
 
 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
4UER
 
 | | 40S-eIF1-eIF1A-eIF3-eIF3j translation initiation complex from Lachancea kluyveri | | Descriptor: | 18S RRNA, EIF1, EIF1A, ... | | Authors: | Aylett, C.H.S, Boehringer, D, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-12-18 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | Structure of a Yeast 40S-Eif1-Eif1A-Eif3-Eif3J Initiation Complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UJC
 
 | | mammalian 80S HCV-IRES initiation complex with eIF5B POST-like state | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Initiation Factor 5B on Hcv-Ires RNA.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4UKD
 
 | | UMP/CMP KINASE FROM SLIME MOLD COMPLEXED WITH ADP, UDP, BERYLLIUM FLUORIDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM DIFLUORIDE, MAGNESIUM ION, ... | | Authors: | Schlichting, I, Reinstein, J. | | Deposit date: | 1997-05-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of active conformations of UMP kinase from Dictyostelium discoideum suggest phosphoryl transfer is associative.
Biochemistry, 36, 1997
|
|
7QEP
 
 | | Cryo-EM structure of the ribosome from Encephalitozoon cuniculi | | Descriptor: | 18S ribosomal RNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Nicholson, D, Ranson, N.A, Melnikov, S.V. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Adaptation to genome decay in the structure of the smallest eukaryotic ribosome
Nat Commun, 13, 2022
|
|
3ZCX
 
 | |
3X2S
 
 | | Crystal structure of pyrene-conjugated adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujii, A, Sekiguchi, Y, Matsumura, H, Inoue, T, Chung, W.-S, Hirota, S, Matsuo, T. | | Deposit date: | 2014-12-31 | | Release date: | 2015-04-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Excimer Emission Properties on Pyrene-Labeled Protein Surface: Correlation between Emission Spectra, Ring Stacking Modes, and Flexibilities of Pyrene Probes.
Bioconjug.Chem., 26, 2015
|
|
6WDR
 
 | | Subunit joining exposes nascent pre-40S rRNA for processing and quality control | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S10-A, ... | | Authors: | Rai, J, Parker, M.D, Huang, H, Choy, S, Ghalei, H, Johnson, M.C, Karbstein, K, Stroupe, M.E. | | Deposit date: | 2020-04-01 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An open interface in the pre-80S ribosome coordinated by ribosome assembly factors Tsr1 and Dim1 enables temporal regulation of Fap7.
Rna, 27, 2021
|
|
8Q2B
 
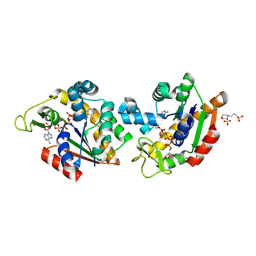 | | E. coli Adenylate Kinase variant D158A (AK D158A) showing significant changes to the stacking of catalytic arginine side chains | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Sauer, U.H, Wolf-Watz, M, Nam, K. | | Deposit date: | 2023-08-01 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidating Dynamics of Adenylate Kinase from Enzyme Opening to Ligand Release.
J.Chem.Inf.Model., 64, 2024
|
|
4V8M
 
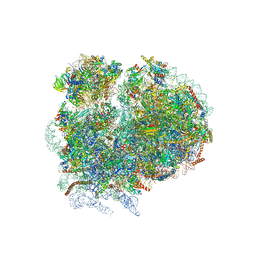 | | High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome | | Descriptor: | 18S RRNA OF THE SMALL RIBOSOMAL SUBUNIT, 40S RIBOSOMAL PROTEIN S10, PUTATIVE, ... | | Authors: | Hashem, Y, des Georges, A, Fu, J, Buss, S.N, Jossinet, F, Jobe, A, Zhang, Q, Liao, H.Y, Grassucci, R.A, Bajaj, C, Westhof, E, Madison-Antenucci, S, Frank, J. | | Deposit date: | 2012-12-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.57 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure of the Trypanosoma Brucei Ribosome.
Nature, 494, 2013
|
|
4WEO
 
 | |
4V5O
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH INITIATION FACTOR 1. | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S12, 40S RIBOSOMAL PROTEIN S3A, ... | | Authors: | Rabl, J, Leibundgut, M, Ataide, S.F, Haag, A, Ban, N. | | Deposit date: | 2010-11-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1.
Science, 331, 2011
|
|
7MHT
 
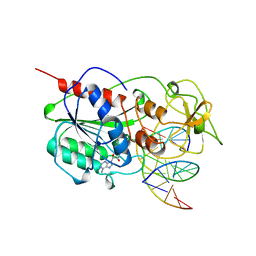 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*AP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-05 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
8Q87
 
 | | Structure of the G. gallus 80S rotated ribosome in complex with eEF2 and SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Nurullina, L, Jenner, L, Yusupov, M. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structure of the inactive ribosome complex accumulated in chick embryo cells in cold-stress conditions.
Febs Lett., 598, 2024
|
|
