3NSC
 
 | | C500S MUTANT OF CueO BOUND TO Cu(II) | | Descriptor: | ACETATE ION, Blue copper oxidase cueO, COPPER (II) ION, ... | | Authors: | Roberts, S.A, Montfort, W.R, Singh, S.K. | | Deposit date: | 2010-07-01 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
3NTP
 
 | | Human Pin1 complexed with reduced amide inhibitor | | Descriptor: | (2R)-2-(acetylamino)-3-[(2S)-2-{[2-(1H-indol-3-yl)ethyl]carbamoyl}pyrrolidin-1-yl]propyl dihydrogen phosphate, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Zhang, Y. | | Deposit date: | 2010-07-05 | | Release date: | 2012-01-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | A reduced-amide inhibitor of Pin1 binds in a conformation resembling a twisted-amide transition state.
Biochemistry, 50, 2011
|
|
5MJG
 
 | | Single-shot pink beam serial crystallography: Thaumatin | | Descriptor: | S,R MESO-TARTARIC ACID, SODIUM ION, Thaumatin-1 | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-shot pink beam serial crystallography: Thaumatin
To Be Published
|
|
5MJQ
 
 | | Single-shot pink beam serial crystallography: Phycocyanin (One chip) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V, Sarrou, I. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
3NZW
 
 | | Crystal structure of the yeast 20S proteasome in complex with 2b | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Gallastegui, N, Marechal, X, Le Ravalec, V, Basse, N, Richy, N, Genin, E, Huber, R, Moroder, M, Vidal, V, Reboud-Ravaux, M. | | Deposit date: | 2010-07-17 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 20S proteasome inhibition: designing noncovalent linear peptide mimics of the natural product TMC-95A.
Chemmedchem, 5, 2010
|
|
3J7Q
 
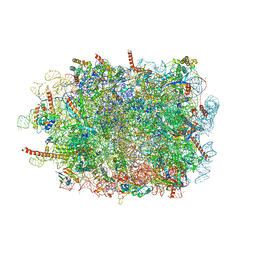 | | Structure of the idle mammalian ribosome-Sec61 complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
5MJU
 
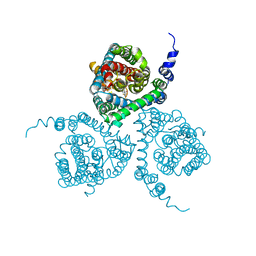 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with the competititve inhibitor TFB-TBOA and the allosteric inhibitor UCPH101 | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1 | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-12-01 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
4HP2
 
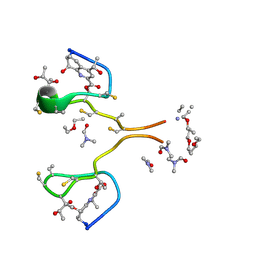 | | Invariom refinement of a new dimeric monoclinic 2 solvate of thiostrepton at 0.64 angstrom resolution | | Descriptor: | DIMETHYLFORMAMIDE, Thiostrepton, diethyl ether | | Authors: | Proepper, K, Holstein, J.J, Huebschle, C.B, Bond, C.S, Dittrich, B. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-02 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (0.64 Å) | | Cite: | Invariom refinement of a new monoclinic solvate of thiostrepton at 0.64 angstrom resolution.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4QNJ
 
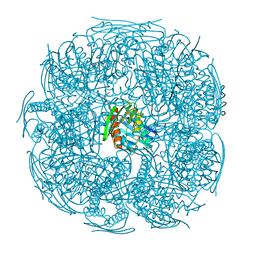 | | The structure of wt A. thaliana IGPD2 in complex with Mn2+ and formate at 1.3A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2014-06-18 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase Is Controlled by Switching Mn(II) Coordination.
Structure, 23, 2015
|
|
5MS3
 
 | | Kallikrein-related peptidase 8 calcium complex | | Descriptor: | CALCIUM ION, Kallikrein-8 | | Authors: | Debela, M, Magdolen, V, Skala, W, Bode, W, Brandstetter, H, Goettig, P. | | Deposit date: | 2016-12-30 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specificity profiles and antagonistic Ca2+ and Zn2+ regulation of human KLK8/neuropsin activity by modules identified in crystal structures
To Be Published
|
|
1W6X
 
 | | SH3 domain of p40phox, component of the NADPH oxidase | | Descriptor: | NEUTROPHIL CYTOSOL FACTOR 4 | | Authors: | Massenet, C, Chenavas, S, Cohen-Addad, C, Dagher, M.-C, Brandolin, G, Pebay-Peyroula, E, Fieschi, F. | | Deposit date: | 2004-08-24 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of P47Phox C-Terminus Phosphorylation on Binding Interactions with P40Phox and P67Phox: Structural and Functional Comparison of P40Phox P67Phox SH3 Domains
J.Biol.Chem., 280, 2005
|
|
3JBR
 
 | | Cryo-EM structure of the rabbit voltage-gated calcium channel Cav1.1 complex at 4.2 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Voltage-dependent L-type calcium channel subunit alpha-1S, ... | | Authors: | Wu, J.P, Yan, Z, Yan, N. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 complex
Science, 350, 2015
|
|
7APE
 
 | | Crystal structure of LpqY from Mycobacterium thermoresistible in complex with trehalose | | Descriptor: | Lipoprotein (Sugar-binding) lpqY, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Furze, C.M, Guy, C.M, Angula, J, Cameron, A.D, Fullam, E. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of trehalose recognition by the mycobacterial LpqY-SugABC transporter.
J.Biol.Chem., 296, 2021
|
|
3JC7
 
 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | Descriptor: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3JCT
 
 | | Cryo-em structure of eukaryotic pre-60S ribosomal subunits | | Descriptor: | 60S ribosomal protein L11-A, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Wu, S, Kumcuoglu, B, Yan, K.G, Brown, H, Zhang, Y.X, Tan, D, Gamalinda, M, Yuan, Y, Li, Z.F, Jakovljevic, J, Ma, C.Y, Lei, J.L, Dong, M.Q, Woolford Jr, J.L, Gao, N. | | Deposit date: | 2016-03-09 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes
Nature, 534, 2016
|
|
4Q4Z
 
 | | Thermus thermophilus RNA polymerase de novo transcription initiation complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ADENOSINE-5'-TRIPHOSPHATE, DNA (25-MER), ... | | Authors: | Murakami, K.S. | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme.
J.Biol.Chem., 289, 2014
|
|
1VRP
 
 | | The 2.1 Structure of T. californica Creatine Kinase Complexed with the Transition-State Analogue Complex, ADP-Mg 2+ /NO3-/Creatine | | Descriptor: | (DIAMINOMETHYL-METHYL-AMINO)-ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, Creatine Kinase, ... | | Authors: | Lahiri, S.D, Wang, P.F, Babbitt, P.C, McLeish, M.J, Kenyon, G.L, Allen, K.N. | | Deposit date: | 2005-04-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Structure of Torpedo californica Creatine Kinase Complexed with the ADP-Mg(2+)-NO3(-)-Creatine Transition-State Analogue Complex
Biochemistry, 41, 2002
|
|
3JA8
 
 | | Cryo-EM structure of the MCM2-7 double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Minichromosome Maintenance 2, Minichromosome Maintenance 3, ... | | Authors: | Li, N, Zhai, Y, Zhang, Y, Li, W, Yang, M, Lei, J, Tye, B.K, Gao, N. | | Deposit date: | 2015-05-09 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the eukaryotic MCM complex at 3.8 angstrom
Nature, 524, 2015
|
|
3JAM
 
 | | CryoEM structure of 40S-eIF1A-eIF1 complex from yeast | | Descriptor: | 18S rRNA, MAGNESIUM ION, RACK1, ... | | Authors: | Llacer, J.L, Hussain, T, Ramakrishnan, V. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex.
Mol.Cell, 59, 2015
|
|
5MXN
 
 | | Atomic model of the VipA/VipB/Hcp, the type six secretion system non-contractile sheath-tube of Vibrio cholerae from cryo-EM | | Descriptor: | Haemolysin co-regulated protein, Type VI secretion protein | | Authors: | Wang, J, Brackmann, M, Castano-Diez, D, Kudryashev, M, Goldie, K, Maier, T, Stahlberg, H, Basler, M. | | Deposit date: | 2017-01-23 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the extended type VI secretion system sheath-tube complex.
Nat Microbiol, 2, 2017
|
|
3O9M
 
 | | Co-crystallization studies of full length recombinant BChE with cocaine offers insights into cocaine detoxification | | Descriptor: | BENZOIC ACID, Cholinesterase | | Authors: | Asojo, O.A, Ngamelue, M.N, Homma, K, Lockridge, O. | | Deposit date: | 2010-08-04 | | Release date: | 2011-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Cocrystallization studies of full-length recombinant butyrylcholinesterase (BChE) with cocaine.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3JCJ
 
 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2015-12-18 | | Release date: | 2016-03-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
4I3J
 
 | | Structures of PR1 intermediate of photoactive yellow protein E46Q mutant from time-resolved laue crystallography collected AT 14ID APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
3NT0
 
 | |
3J6X
 
 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 5 degree rotation (Class II) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
