5HG1
 
 | | Crystal Structure of Human Hexokinase 2 with cmpd 1, a C-2-substituted glucosamine | | Descriptor: | 2-deoxy-2-{[(2E)-3-(3,4-dichlorophenyl)prop-2-enoyl]amino}-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
8W1L
 
 | |
5LP1
 
 | |
5MVS
 
 | | Crystal structure of Dot1L in complex with adenosine and inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] | | Descriptor: | ADENOSINE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
6I9Y
 
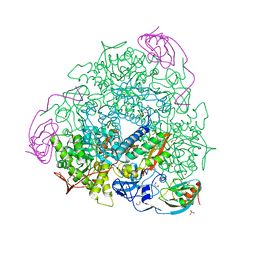 | | The 2.14 A X-ray crystal structure of Sporosarcina pasteurii urease in complex with Au(I) ions | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, HYDROXIDE ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-11-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Inhibition Mechanism of Urease by Au(III) Compounds Unveiled by X-ray Diffraction Analysis.
Acs Med.Chem.Lett., 10, 2019
|
|
7T2X
 
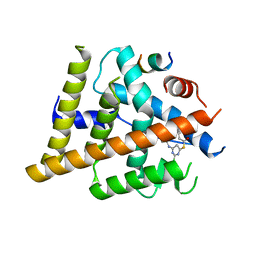 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S in Complex with 2-chloro-4-((4-hydroxybenzyl)amino)-5-phenylthieno[2,3-d]pyrimidin-6-ol and GRIP Peptide | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, S-(2-chloro-6-{[(4-hydroxyphenyl)methyl]amino}pyrimidin-4-yl) phenylethanethioate | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Wilson, T.M, Fanning, S.W. | | Deposit date: | 2021-12-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A New Chemotype of Chemically Tractable Nonsteroidal Estrogens Based on a Thieno[2,3- d ]pyrimidine Core.
Acs Med.Chem.Lett., 13, 2022
|
|
8UN4
 
 | |
6N2J
 
 | | Tetrahydropyridopyrimidines as Covalent Inhibitors of KRAS-G12C | | Descriptor: | 1-{4-[7-(naphthalen-1-yl)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vigers, G.P. | | Deposit date: | 2018-11-13 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Tetrahydropyridopyrimidines as Irreversible Covalent Inhibitors of KRAS-G12C with In Vivo Activity.
ACS Med Chem Lett, 9, 2018
|
|
6K1S
 
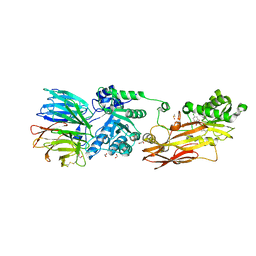 | | Discovery of Potent and Selective Covalent Protein Arginine Methyltransferase (PRMT5) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-[[7-[(2~{R},3~{R},4~{S},5~{R})-5-[(~{R})-(4-chlorophenyl)-oxidanyl-methyl]-3,4-bis(oxidanyl)oxolan-2-yl]pyrrolo[2,3-d]pyrimidin-4-yl]amino]ethanal, DIMETHYL SULFOXIDE, ... | | Authors: | Tong, S, Lin, H. | | Deposit date: | 2019-05-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent and Selective Covalent Protein Arginine Methyltransferase 5 (PRMT5) Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
5LUU
 
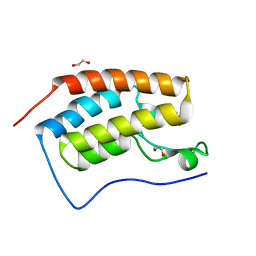 | | Structure of the first bromodomain of BRD4 with a pyrazolo[4,3-c]pyridin fragment | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-phenyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)propan-1-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Knapp, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
5ITD
 
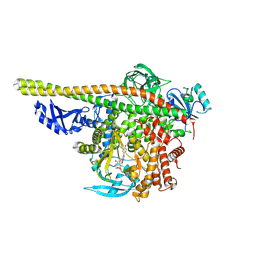 | | Crystal structure of PI3K alpha with PI3K delta inhibitor | | Descriptor: | 5-{4-[3-(4-acetylpiperazine-1-carbonyl)phenyl]quinazolin-6-yl}-2-methoxypyridine-3-carbonitrile, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Discovery and Pharmacological Characterization of Novel Quinazoline-Based PI3K Delta-Selective Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5T92
 
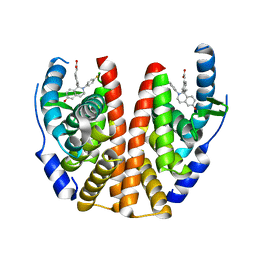 | | ESTROGEN RECEPTOR ALPHA LIGAND BINDING DOMAIN IN COMPLEX WITH (2E)-3-{4-[(1R)-2-(4-fluorophenyl)-6-hydroxy-1-methy l-1,2,3,4- tetrahydroisoquinolin-1-yl]phenyl}prop-2-enoic acid | | Descriptor: | (2E)-3-{4-[(1R)-2-(4-fluorophenyl)-6-hydroxy-1-methyl-1,2,3,4-tetrahydroisoquinolin-1-yl]phenyl}prop-2-enoic acid, Estrogen receptor | | Authors: | Kirby, C, Baird, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of an Acrylic Acid Based Tetrahydroisoquinoline as an Orally Bioavailable Selective Estrogen Receptor Degrader for ER alpha + Breast Cancer.
J. Med. Chem., 60, 2017
|
|
5T97
 
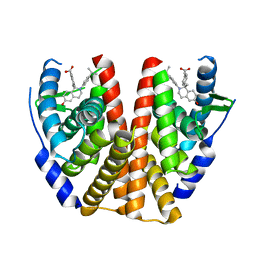 | | ESTROGEN RECEPTOR ALPHA LIGAND BINDING DOMAIN IN COMPLEX WITH (2E)-3-(4-{(1R)-6-hydroxy-1-methyl-2-[4-(propan-2 -yl)phenyl]-1,2,3,4- tetrahydroisoquinolin-1-yl}phenyl)prop-2-enoic acid | | Descriptor: | (2E)-3-(4-{(1R)-6-hydroxy-1-methyl-2-[4-(propan-2-yl)phenyl]-1,2,3,4-tetrahydroisoquinolin-1-yl}phenyl)prop-2-enoic acid, Estrogen receptor | | Authors: | Kirby, C.A, Baird, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of an Acrylic Acid Based Tetrahydroisoquinoline as an Orally Bioavailable Selective Estrogen Receptor Degrader for ER alpha + Breast Cancer.
J. Med. Chem., 60, 2017
|
|
6B34
 
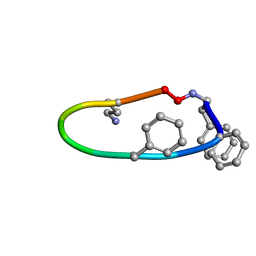 | |
8EC1
 
 | |
8G7F
 
 | | Crystal Structure of FosB from Bacillus cereus with Zinc and 1-hydroxypropylphosphonic acid | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Travis, S, Pang, A.H, Tsodikov, O.V, Garneau-Tsodikova, S, Thompson, M.K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Identification and analysis of small molecule inhibitors of FosB from Staphylococcus aureus.
Rsc Med Chem, 14, 2023
|
|
8G7G
 
 | | Crystal Structure of FosB from Bacillus cereus with Zinc and (1-hydroxy-2-methylpropyl)phosphonic acid | | Descriptor: | FORMIC ACID, MAGNESIUM ION, Metallothiol transferase FosB, ... | | Authors: | Travis, S, Pang, A.H, Tsodikov, O.V, Garneau-Tsodikova, S, Thompson, M.K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Identification and analysis of small molecule inhibitors of FosB from Staphylococcus aureus.
Rsc Med Chem, 14, 2023
|
|
8G7H
 
 | | Crystal Structure of FosB from Bacillus cereus with Zinc and (1-hydroxypropan-2-yl)phosphonic acid | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Travis, S, Pang, A.H, Tsodikov, O.V, Garneau-Tsodikova, S, Thompson, M.K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identification and analysis of small molecule inhibitors of FosB from Staphylococcus aureus.
Rsc Med Chem, 14, 2023
|
|
8G7I
 
 | | Crystal Structure of FosB from Bacillus cereus with Zinc and Sulfate | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Travis, S, Pang, A.H, Tsodikov, O.V, Garneau-Tsodikova, S, Thompson, M.K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification and analysis of small molecule inhibitors of FosB from Staphylococcus aureus.
Rsc Med Chem, 14, 2023
|
|
6AY3
 
 | | CREBBP bromodomain in complex with Cpd16 (5-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1H-indole-3-carboxamide) | | Descriptor: | 1,2-ETHANEDIOL, 5-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1H-indole-3-carboxamide, CREB-binding protein, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
6B35
 
 | |
8E4A
 
 | | Pseudomonas LpxC in complex with LPC-233 | | Descriptor: | 4-(4-cyclopropylbuta-1,3-diyn-1-yl)-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Najeeb, J, Zhou, P. | | Deposit date: | 2022-08-17 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Preclinical safety and efficacy characterization of an LpxC inhibitor against Gram-negative pathogens.
Sci Transl Med, 15, 2023
|
|
6EN7
 
 | |
6XLU
 
 | | Structure of SARS-CoV-2 spike at pH 4.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
5KBR
 
 | | Pak1 in complex with 7-azaindole inhibitor | | Descriptor: | (4-chlorophenyl)-[5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]methanone, Serine/threonine-protein kinase PAK 1 | | Authors: | Ferguson, A. | | Deposit date: | 2016-06-03 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Optimization of Highly Kinase Selective Bis-anilino Pyrimidine PAK1 Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
