2Y3K
 
 | | Structure of segment MVGGVVIA from the amyloid-beta peptide (Ab, residues 35-42), alternate polymorph 1 | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J.P, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-21 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3HC5
 
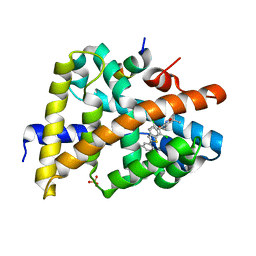 | | FXR with SRC1 and GSK826 | | Descriptor: | 3-(6-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-1-benzothiophen-2-yl)benzoic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | FXR agonist activity of conformationally constrained analogs of GW 4064.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3CSH
 
 | |
3CT1
 
 | |
3CV8
 
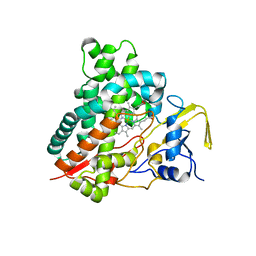 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84F mutant) | | Descriptor: | Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, K, Sugimoto, H, Shinkyo, R, Yamada, M, Ikeda, S, Ikushiro, S, Kamakura, M, Shiro, Y, Sakaki, T. | | Deposit date: | 2008-04-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1
Biochemistry, 47, 2008
|
|
3CWL
 
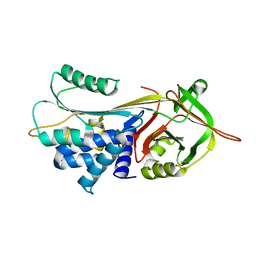 | | Crystal structure of alpha-1-antitrypsin, crystal form B | | Descriptor: | Alpha-1-antitrypsin, CHLORIDE ION | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
3CYF
 
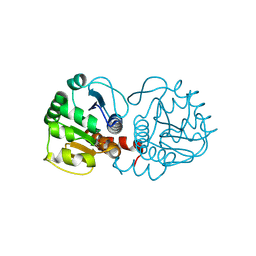 | | Crystal Structure of E18N DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hasim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
3CZ9
 
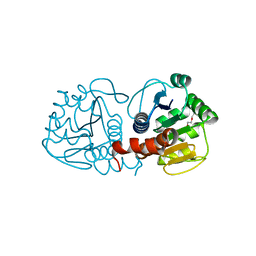 | | Crystal Structure of E18L DJ-1 | | Descriptor: | O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hasim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
3D81
 
 | | Sir2-S-alkylamidate complex crystal structure | | Descriptor: | NAD-dependent deacetylase, S-alkylamidate intermediate, ZINC ION | | Authors: | Hawse, W.F, Hoff, K.G, Fatkins, D, Daines, A, Zubkova, O.V, Schramm, V.L, Zheng, W, Wolberger, C. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into intermediate steps in the Sir2 deacetylation reaction.
Structure, 16, 2008
|
|
1OH5
 
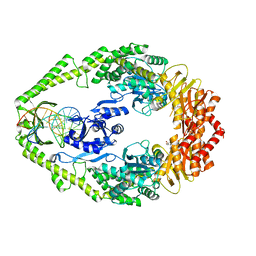 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A C:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*CP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH6
 
 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN A:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*AP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP* CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP* CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
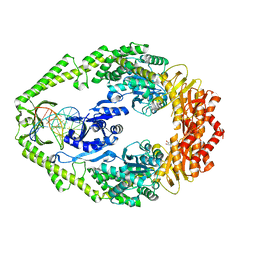
3H47
 
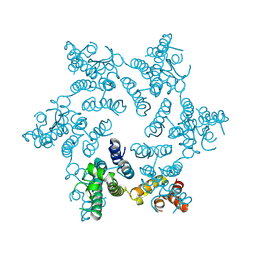 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24 | | Authors: | Pornillos, O. | | Deposit date: | 2009-04-18 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
1OOJ
 
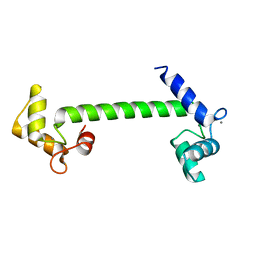 | | Structural genomics of Caenorhabditis elegans : Calmodulin | | Descriptor: | CALCIUM ION, Calmodulin CMD-1 | | Authors: | Symersky, J, Lin, G, Li, S, Qiu, S, Luan, C.-H, Luo, D, Tsao, J, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural genomics of caenorhabditis elegans: crystal structure of calmodulin.
Proteins, 53, 2003
|
|
1OH8
 
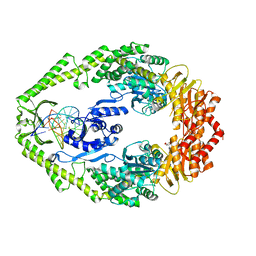 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN UNPAIRED THYMIDINE | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP* TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*CP*TP*TP*GP*GP*CP* AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OT7
 
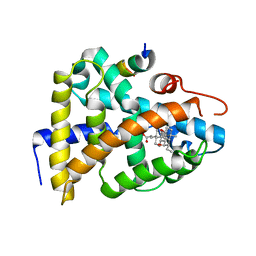 | | Structural Basis for 3-deoxy-CDCA Binding and Activation of FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile Acid Receptor, ISO-URSODEOXYCHOLIC ACID, ... | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1OH7
 
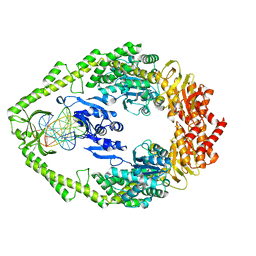 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A G:G MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*GP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OZF
 
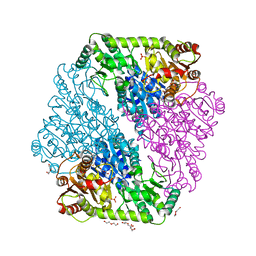 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactors | | Descriptor: | Acetolactate synthase, catabolic, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | Deposit date: | 2003-04-09 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
3GUH
 
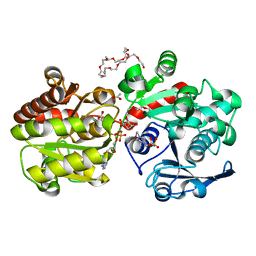 | | Crystal Structure of Wild-type E.coli GS in complex with ADP and DGM | | Descriptor: | (2R)-2-hydroxy-3-[4-(2-hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid, 1,5-anhydro-D-glucitol, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Sheng, F, Geiger, J. | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The Crystal Structures of the Open and Catalytically Competent Closed Conformation of Escherichia coli Glycogen Synthase.
J.Biol.Chem., 284, 2009
|
|
3CZA
 
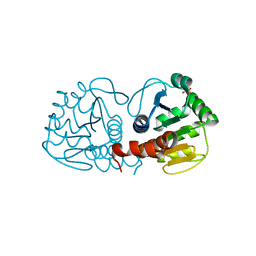 | | Crystal Structure of E18D DJ-1 | | Descriptor: | MALONIC ACID, Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hashim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
1P7H
 
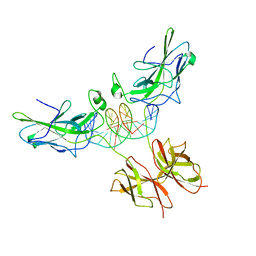 | | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kB element | | Descriptor: | 5'-D(*AP*AP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*A)-3', 5'-D(*TP*TP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*A)-3', Nuclear factor of activated T-cells, ... | | Authors: | Giffin, M.J, Stroud, J.C, Bates, D.L, von Koenig, K.D, Hardin, J, Chen, L. | | Deposit date: | 2003-05-01 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element
Nat.Struct.Biol., 10, 2003
|
|
3GJX
 
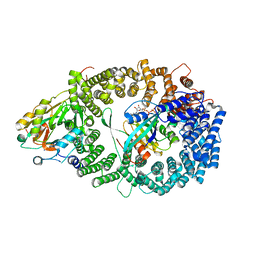 | | Crystal Structure of the Nuclear Export Complex CRM1-Snurportin1-RanGTP | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Monecke, T, Guettler, T, Neumann, P, Dickmanns, A, Goerlich, D, Ficner, R. | | Deposit date: | 2009-03-09 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Nuclear Export Receptor CRM1 in Complex with Snurportin1 and RanGTP.
Science, 2009
|
|
1OZH
 
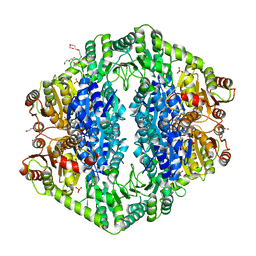 | | The crystal structure of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate. | | Descriptor: | 2-HYDROXYETHYL DIHYDROTHIACHROME DIPHOSPHATE, Acetolactate synthase, catabolic, ... | | Authors: | Pang, S.S, Duggleby, R.G, Schowen, R.L, Guddat, L.W. | | Deposit date: | 2003-04-09 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structures of Klebsiella pneumoniae Acetolactate Synthase with Enzyme-bound Cofactor and with an Unusual Intermediate.
J.Biol.Chem., 279, 2004
|
|
1Q81
 
 | | Crystal Structure of minihelix with 3' puromycin bound to A-site of the 50S ribosomal subunit. | | Descriptor: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Insights into Peptide Bond Formation
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3GQ7
 
 | |
3GKE
 
 | | Crystal Structure of Dicamba Monooxygenase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DdmC, ... | | Authors: | Wilson, M.A, Dumitru, R, Jiang, W.Z, Weeks, D.P. | | Deposit date: | 2009-03-10 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dicamba monooxygenase: a Rieske nonheme oxygenase that catalyzes oxidative demethylation.
J.Mol.Biol., 392, 2009
|
|
