8R5C
 
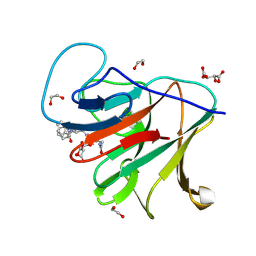 | | Crystal structure of human TRIM7 PRYSPRY domain bound to (2-(1-oxoisoindolin-2-yl)-3-phenylpropanoyl)-L-glutamine | | Descriptor: | (2~{S})-5-azanyl-5-oxidanylidene-2-[[(2~{S})-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)-3-phenyl-propanoyl]amino]pentanoic acid, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, ... | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
3S2V
 
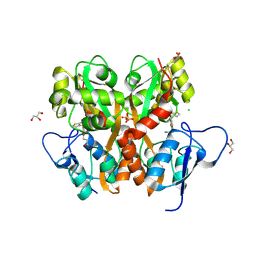 | | Crystal Structure of the Ligand Binding Domain of GluK1 in Complex with an Antagonist (S)-1-(2'-Amino-2'-carboxyethyl)-3-[(2-carboxythien-3-yl)methyl]thieno[3,4-d]pyrimidin-2,4-dione at 2.5 A Resolution | | Descriptor: | (S)-1-(2'-AMINO-2'-CARBOXYETHYL)-3-[(2-CARBOXYTHIEN-3-YL)METHYL]THIENO[3,4-D]PYRIMIDIN-2,4-DIONE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective kainate receptor (GluK1) ligands structurally based upon 1H-cyclopentapyrimidin-2,4(1H,3H)-dione: synthesis, molecular modeling, and pharmacological and biostructural characterization.
J.Med.Chem., 54, 2011
|
|
1L5G
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN AVB3 IN COMPLEX WITH AN ARG-GLY-ASP LIGAND | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.-P, Stehle, T, Zhang, R, Joachimiak, A, Frech, M, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2002-03-06 | | Release date: | 2002-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand.
Science, 296, 2002
|
|
2I1P
 
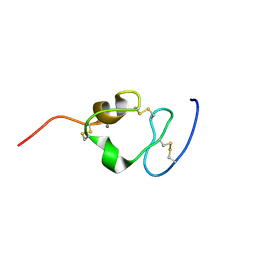 | | Solution structure of the twelfth cysteine-rich ligand-binding repeat in rat megalin | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 2 | | Authors: | Wolf, C.A, Dancea, F, Shi, M, Bade-Noskova, V, Rueterjans, H, Kerjaschki, D, Luecke, C. | | Deposit date: | 2006-08-14 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the twelfth cysteine-rich ligand-binding repeat in rat megalin.
J.Biomol.Nmr, 37, 2007
|
|
7QEZ
 
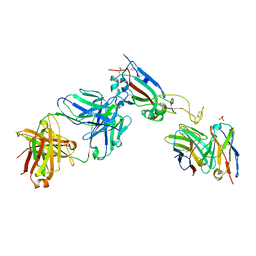 | |
8PD6
 
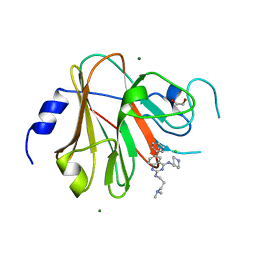 | | Crystal structure of the TRIM58 PRY-SPRY domain in complex with TRIM-473 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase TRIM58, ... | | Authors: | Renatus, M, Hoegenauer, K, Schroeder, M. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Ligands for TRIM58, a Novel Tissue-Selective E3 Ligase.
Acs Med.Chem.Lett., 14, 2023
|
|
8PD4
 
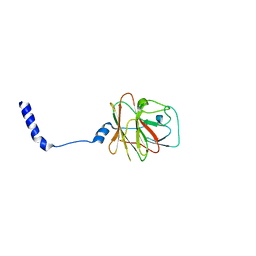 | |
1L2J
 
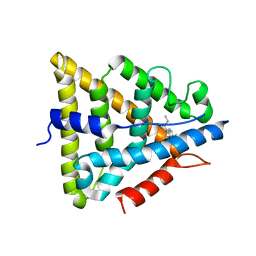 | | Human Estrogen Receptor beta Ligand-binding Domain in Complex with (R,R)-5,11-cis-diethyl-5,6,11,12-tetrahydrochrysene-2,8-diol | | Descriptor: | (R,R)-5,11-CIS-DIETHYL-5,6,11,12-TETRAHYDROCHRYSENE-2,8-DIOL, ESTROGEN RECEPTOR BETA | | Authors: | Shiau, A.K, Barstad, D, Radek, J.T, Meyers, M.J, Nettles, K.W, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Agard, D.A, Greene, G.L. | | Deposit date: | 2002-02-21 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism.
Nat.Struct.Biol., 9, 2002
|
|
5XUA
 
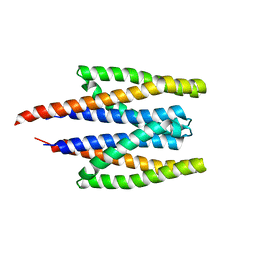 | |
2QEJ
 
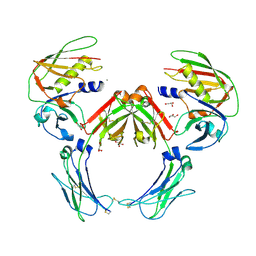 | | Crystal structure of a Staphylococcus aureus protein (SSL7) in complex with Fc of human IgA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Ramsland, P.A, Willoughby, N, Trist, H.M, Farrugia, W, Hogarth, P.M, Fraser, J.D, Wines, B.D. | | Deposit date: | 2007-06-26 | | Release date: | 2007-09-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7TTZ
 
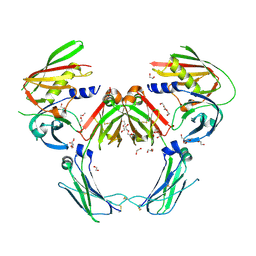 | | Heterodimeric IgA Fc in complex with Staphylococcus aureus protein SSL7 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Boulanger, M.J, Verstraete, M, Heinkel, F, Escobar, E, Dixit, S, Von Kreudenstein, T.S. | | Deposit date: | 2022-02-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Engineering a pure and stable heterodimeric IgA for the development of multispecific therapeutics.
Mabs, 14, 2022
|
|
8R5D
 
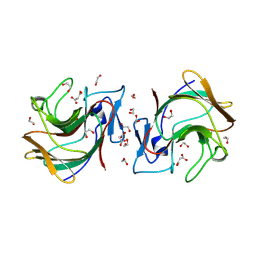 | | Crystal structure of human TRIM7 PRYSPRY domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, MALONIC ACID | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
6KYP
 
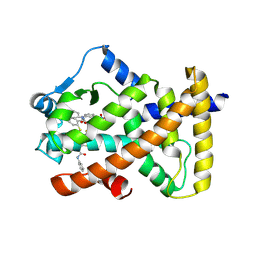 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-clofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-(4-chloranylphenoxy)-2-methyl-propanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-09-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L37
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L36
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L38
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-gemfibrozil co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.761 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
5DGW
 
 | | Crystal Structure of HIV-1 Protease Inhibitor GRL-105-11A Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand | | Descriptor: | (3R,3aS,4S,7aS)-3-(ethylamino)hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Pol protein, ... | | Authors: | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2015-08-28 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
6LX4
 
 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX9
 
 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX5
 
 | | X-ray structure of human PPARalpha ligand binding domain-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
8R6B
 
 | |
6QTN
 
 | | Tubulin-cyclostreptin complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Balaguer, F.d.A, Muehlethaler, T, Estevez-Gallego, J, Calvo, E, Gimenez-Abian, J.F, Risinger, A.L, Sorensen, E.J, Vanderwal, C.D, Altmann, K.-H, Mooberry, S.L, Steinmetz, M.O, Oliva, M.A, Prota, A.E, Diaz, J.F. | | Deposit date: | 2019-02-25 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Cyclostreptin-Tubulin Adduct: Implications for Tubulin Activation by Taxane-Site Ligands.
Int J Mol Sci, 20, 2019
|
|
1LAH
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | L-ornithine, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
6MD4
 
 | |
6MD1
 
 | |
