1JY6
 
 | |
3JCI
 
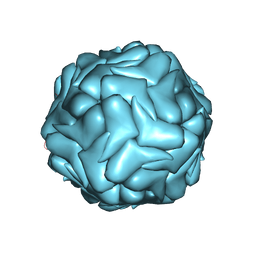 | | 2.9 Angstrom Resolution Cryo-EM 3-D Reconstruction of Close-packed PCV2 Virus-like Particles | | Descriptor: | Capsid protein | | Authors: | Liu, Z, Guo, F, Wang, F, Li, T.C, Jiang, W. | | Deposit date: | 2015-12-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | 2.9 angstrom Resolution Cryo-EM 3D Reconstruction of Close-Packed Virus Particles.
Structure, 24, 2016
|
|
8ATV
 
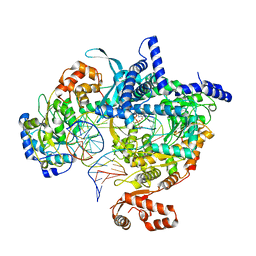 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 5-mer RNA, pppGpGpApApA (IC5) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATW
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 6-mer RNA, pppGpGpApApApU (IC6) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8ATT
 
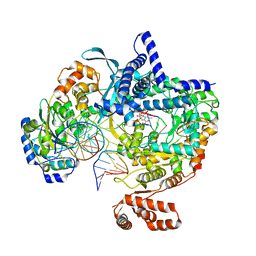 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 4-mer RNA, pppGpGpUpA (IC4) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
6NLB
 
 | | Crystal structure of de novo designed metal-controlled dimer of mutant B1 immunoglobulin-binding domain of Streptococcal Protein G (L12H, E15V, T16L, T18I, V29H, Y33H, N37L)-apo | | Descriptor: | Immunoglobulin G-binding protein G, MAGNESIUM ION | | Authors: | Maniaci, B, Stec, B, Huxford, T. | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of High-Affinity Metal-Controlled Protein Dimers.
Biochemistry, 58, 2019
|
|
6NL8
 
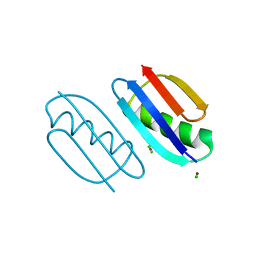 | | Crystal structure of de novo designed metal-controlled dimer of mutant B1 immunoglobulin-binding domain of Streptococcal Protein G (L12H, T16L, V29H, Y33H, N37L)-zinc | | Descriptor: | CHLORIDE ION, Immunoglobulin G-binding protein G, ZINC ION | | Authors: | Maniaci, B, Stec, B, Huxford, T. | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of High-Affinity Metal-Controlled Protein Dimers.
Biochemistry, 58, 2019
|
|
6NZU
 
 | | Structure of the human frataxin-bound iron-sulfur cluster assembly complex | | Descriptor: | Acyl carrier protein, Cysteine desulfurase, mitochondrial, ... | | Authors: | Fox, N.G, Yu, X, Xidong, F, Alain, M, Joseph, N, Claire, S.D, Christine, B, Han, S, Yue, W.W. | | Deposit date: | 2019-02-14 | | Release date: | 2019-05-22 | | Last modified: | 2019-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism.
Nat Commun, 10, 2019
|
|
5WLW
 
 | |
4OIO
 
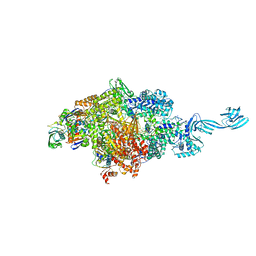 | | Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
1JY4
 
 | |
4NDL
 
 | | Computational design and experimental verification of a symmetric homodimer | | Descriptor: | ENH-c2b, computational designed homodimer | | Authors: | Mou, Y, Huang, P.S, Hsu, F.C, Huang, S.J, Mayo, S.L. | | Deposit date: | 2013-10-26 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Computational design and experimental verification of a symmetric protein homodimer.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1QCZ
 
 | |
1C3T
 
 | |
7OMM
 
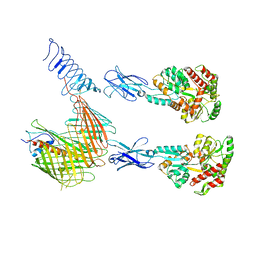 | | Cryo-EM structure of N. gonorhoeae LptDE in complex with ProMacrobodies (MBPs have not been built de novo) | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ProMacrobody 21,Maltodextrin-binding protein, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
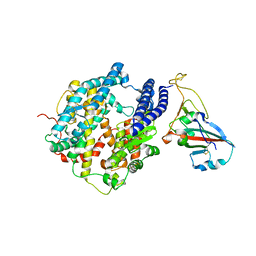
7P19
 
 | |
1D3G
 
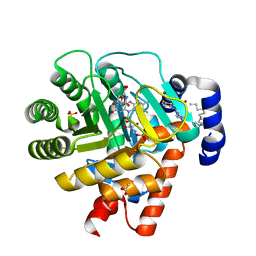 | | HUMAN DIHYDROOROTATE DEHYDROGENASE COMPLEXED WITH BREQUINAR ANALOG | | Descriptor: | 2-BIPHENYL-4-YL-6-FLUORO-3-METHYL-QUINOLINE-4-CARBOXYLIC ACID, ACETATE ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Liu, S, Neidhardt, E.A, Grossman, T.H, Ocain, T, Clardy, J. | | Deposit date: | 1999-09-29 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents.
Structure Fold.Des., 8, 2000
|
|
1D3H
 
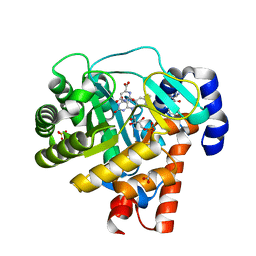 | | HUMAN DIHYDROOROTATE DEHYDROGENASE COMPLEXED WITH ANTIPROLIFERATIVE AGENT A771726 | | Descriptor: | (2Z)-2-cyano-3-hydroxy-N-[4-(trifluoromethyl)phenyl]but-2-enamide, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Liu, S, Neidhardt, E.A, Grossman, T.H, Ocain, T, Clardy, J. | | Deposit date: | 1999-09-29 | | Release date: | 2000-08-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents.
Structure Fold.Des., 8, 2000
|
|
1F1O
 
 | | STRUCTURAL STUDIES OF ADENYLOSUCCINATE LYASES | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T. | | Deposit date: | 2000-05-19 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
1GSO
 
 | |
3LTB
 
 | | X-ray structure of a non-biological ATP binding protein determined in the presence of 10 mM ATP at 2.6 A after 3 weeks of incubation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
5BYO
 
 | |
2N6B
 
 | |
5DI5
 
 | | beta1 t801 loop variant in P3221 | | Descriptor: | beta1 t801 loop variant | | Authors: | MacDonald, J.T, Kabasakal, B.V, Murray, J.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KUX
 
 | |
