9B0B
 
 | |
9B03
 
 | | INF2 in the Middle of F-Actin (Up state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
9B00
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with berberine analog of chloramphenicol CAM-BER, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.80A resolution | | Descriptor: | 13-(2-{[(1R,2R)-1,3-dihydroxy-1-(4-nitrophenyl)propan-2-yl]amino}-2-oxoethyl)-9,10-dimethoxy-5,6-dihydro-2H-[1,3]dioxolo[4,5-g]isoquinolino[3,2-a]isoquinolin-7-ium, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Batool, Z, Pavlova, J.A, Paranjpe, M.N, Tereshchenkov, A.G, Lukianov, D.A, Osterman, I.A, Bogdanov, A.A, Sumbatyan, N.V, Polikanov, Y.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Berberine analog of chloramphenicol exhibits a distinct mode of action and unveils ribosome plasticity.
Structure, 32, 2024
|
|
9AZZ
 
 | |
9AZX
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with NDPr | | Descriptor: | Non-structural protein 3, {(2R,3S,4R,5R)-5-[(8S)-4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl]-5-cyano-3,4-dihydroxyoxolan-2-yl}methyl [(2R,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate | | Authors: | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | GS-441524-Diphosphate-Ribose Derivatives as Nanomolar Binders and Fluorescence Polarization Tracers for SARS-CoV-2 and Other Viral Macrodomains.
Acs Chem.Biol., 19, 2024
|
|
9AZQ
 
 | | INF2 at the Barbed End of F-Actin with Incoming Actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
9AZP
 
 | | INF2 at the Barbed End of F-Actin with Incoming Profilin-Actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
9AZO
 
 | | Crystal structure of CHMS Dehydrogenase PmdC from Comamonas testosteroni bound to cofactor NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PmdC, SULFATE ION | | Authors: | Rodrigues, A.V, Moriarty, N.W, Pereira, J.H, Adams, P.D. | | Deposit date: | 2024-03-11 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Characterization of lignin-degrading enzyme PmdC, which catalyzes a key step in the synthesis of polymer precursor 2-pyrone-4,6-dicarboxylic acid.
J.Biol.Chem., 300, 2024
|
|
9AZJ
 
 | |
9AZI
 
 | |
9AZB
 
 | | Crystal structure of LolTv5 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-11 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
9AZA
 
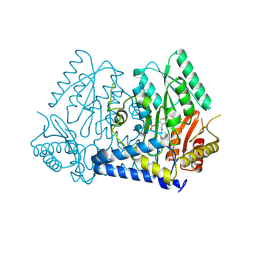 | | Crystal structure of LolTv4 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
9AZ9
 
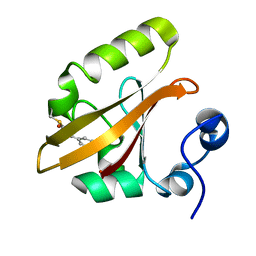 | | Chloride Sites in Photoactive Yellow Protein (Chloride-Free Reference Structure) | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Dyda, F, Schotte, F, Anfinrud, P, Cho, H.S. | | Deposit date: | 2024-03-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Watching a signaling protein function: What has been learned over four decades of time-resolved studies of photoactive yellow protein.
Struct Dyn., 11, 2024
|
|
9AZ7
 
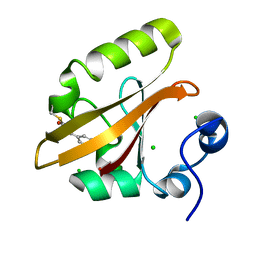 | | Chloride Sites in Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, CHLORIDE ION, Photoactive yellow protein | | Authors: | Dyda, F, Schotte, F, Anfinrud, P, Cho, H.S. | | Deposit date: | 2024-03-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Watching a signaling protein function: What has been learned over four decades of time-resolved studies of photoactive yellow protein.
Struct Dyn., 11, 2024
|
|
9AZ4
 
 | | INF2 at the Barbed End of F-Actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-10 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
9AYY
 
 | |
9AYX
 
 | |
9AYW
 
 | |
9AYM
 
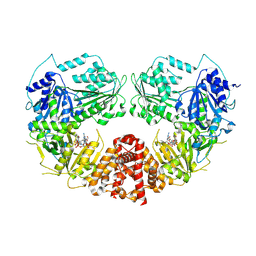 | | Cryo-EM Structure of E.coli produced recombinant N-acetyltransferase 10 (NAT10) in complex with cytidine-acetone-CoA bisubstrate probe | | Descriptor: | RNA cytidine acetyltransferase, [[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{S})-4-[[3-[2-[3-[1-[(2~{R},3~{R},4~{R},5~{S})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]sulfanyl-2-oxidanylidene-propyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Zhou, M, Marmorstein, R. | | Deposit date: | 2024-03-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Molecular Basis for RNA Cytidine Acetylation by NAT10.
Biorxiv, 2024
|
|
9AYL
 
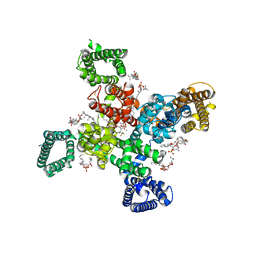 | | Cryo-EM structure of human Cav3.2 with ACT-709478 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYK
 
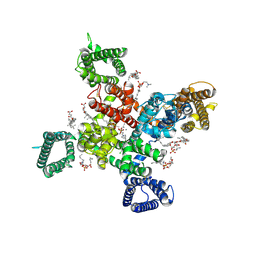 | | Cryo-EM structure of human Cav3.2 with ML218 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,5-dichloro-N-{[(1R,5S,6r)-3-(3,3-dimethylbutyl)-3-azabicyclo[3.1.0]hexan-6-yl]methyl}benzamide, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYJ
 
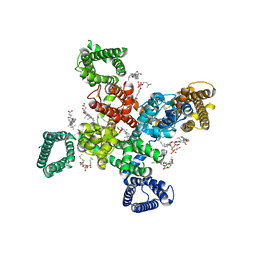 | | Cryo-EM structure of human Cav3.2 with TTA-P2 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,5-dichloro-N-[(1-{[(4S)-2,2-dimethyloxan-4-yl]methyl}-4-fluoropiperidin-4-yl)methyl]benzamide, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYH
 
 | | Cryo-EM structure of human Cav3.2 with TTA-A2 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(4-cyclopropylphenyl)-N-{(1R)-1-[5-(2,2,2-trifluoroethoxy)pyridin-2-yl]ethyl}acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYG
 
 | | Cryo-EM structure of apo state human Cav3.2 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYF
 
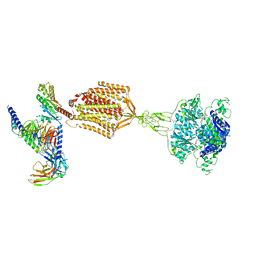 | | Structure of human calcium-sensing receptor in complex with Gi1 (miniGi1) protein in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
