8UQE
 
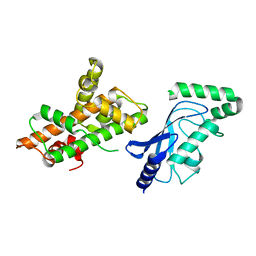 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 26-residue linker (RING not modeled in density) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.562 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQA
 
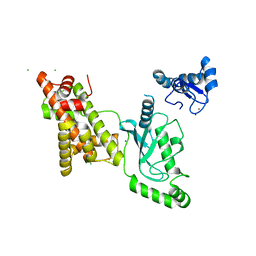 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 12-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, SODIUM ION, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQB
 
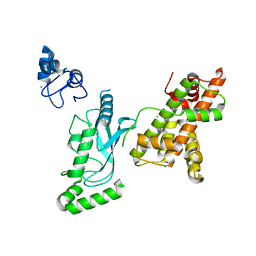 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 1) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQ9
 
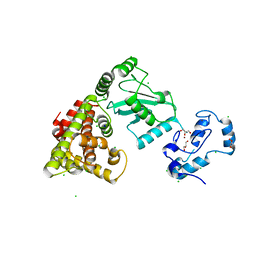 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 4-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQ8
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 2-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQD
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (condition 2. RING not modeled in density) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.893 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQC
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 2) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TIM
 
 | |
8SW0
 
 | | Puromycin sensitive aminopeptidase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Puromycin-sensitive aminopeptidase, ... | | Authors: | Rodgers, D.W, Sampath, S. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of puromycin-sensitive aminopeptidase and polyglutamine binding.
Plos One, 18, 2023
|
|
8VIJ
 
 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin Y34F C51S C71S variant (cyanomet) | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lecomte, J.T.J, Schlessman, J.L, Martinez, J.E, Schultz, T.D, Siegler, M.A, DelCampo, M, Le Magueres, P. | | Deposit date: | 2024-01-04 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin Y34F C51S C71S variant in the cyanomet state
To be published
|
|
8VK2
 
 | | X-ray crystal structure of human IgE 4C8 Fab | | Descriptor: | IgE 4C8 heavy chain, IgE 4C8 light chain | | Authors: | Khatri, K, Ball, A, Smith, S.A, Champan, M.D, Pomes, A, Chruszcz, M. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Structural analysis of human IgE monoclonal antibody epitopes on dust mite allergen Der p 2.
J.Allergy Clin.Immunol., 154, 2024
|
|
8VK1
 
 | | X-ray crystal structure of human IgE 4C8 Fab complex with Der p 2.0103 | | Descriptor: | Der p 2 variant 3, IgE 4C8 heavy chain, IgE 4C8 light chain | | Authors: | Khatri, K, Ball, A, Smith, S.A, Champan, M.D, Pomes, A, Chruszcz, M. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural analysis of human IgE monoclonal antibody epitopes on dust mite allergen Der p 2.
J.Allergy Clin.Immunol., 154, 2024
|
|
7ENN
 
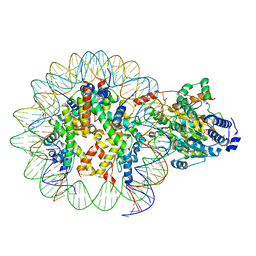 | | The structure of ALC1 bound to the nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromodomain-helicase-DNA-binding protein 1-like, ... | | Authors: | Chen, Z.C, Chen, K.J, Wang, L. | | Deposit date: | 2021-04-18 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome.
Nat Commun, 12, 2021
|
|
7EPC
 
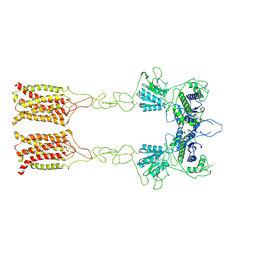 | | Cryo-EM structure of inactive mGlu7 homodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPB
 
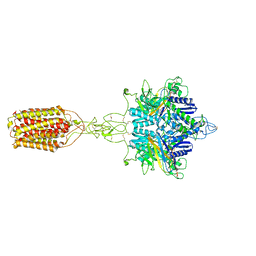 | | Cryo-EM structure of LY354740-bound mGlu2 homodimer | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, Anti-RON nanobody, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPA
 
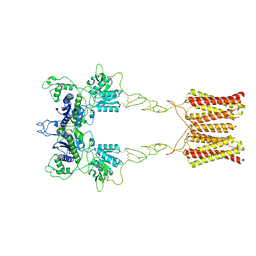 | | Cryo-EM structure of inactive mGlu2 homodimer | | Descriptor: | Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPD
 
 | | Cryo-EM structure of inactive mGlu2-7 heterodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7, Metabotropic glutamate receptor 2,Peptidylprolyl isomerase | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPU
 
 | | Crystal structure of HsALC1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromodomain-helicase-DNA-binding protein 1-like, MAGNESIUM ION, ... | | Authors: | Wang, L, Chen, K.J. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome.
Nat Commun, 12, 2021
|
|
7EXP
 
 | | Crystal structure of zebrafish TRAP1 with AMPPNP and MitoQ | | Descriptor: | 2,3-dimethoxy-5-methyl-6-[10-(triphenyl-$l^{5}-phosphanyl)decyl]cyclohexa-2,5-diene-1,4-dione, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Lee, H, Yoon, N.G, Kang, B.H, Lee, C. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Mitoquinone Inactivates Mitochondrial Chaperone TRAP1 by Blocking the Client Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
7F50
 
 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with AMPPnP | | Descriptor: | CHLORIDE ION, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | Authors: | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
7EST
 
 | |
7EDZ
 
 | | Crystal Structure of human PPCS in complex with P-HoPan and AMPPNP | | Descriptor: | 4-[[(2R)-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butanoyl]amino]butanoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sharma, N, Mostert, K.J, Strauss, E, Anand, R. | | Deposit date: | 2021-03-17 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Coenzyme A Level Modulator Hopantenate (HoPan) Inhibits Phosphopantotenoylcysteine Synthetase Activity.
Acs Chem.Biol., 16, 2021
|
|
7DRN
 
 | | Structure of ATP-grasp ligase PsnB complexed with precursor peptide PsnA2 and AMPPNP | | Descriptor: | ATP-grasp domain-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PsnA214-38, ... | | Authors: | Song, I, Yu, J, Song, W, Kim, S. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB.
Nat.Chem.Biol., 17, 2021
|
|
2FRZ
 
 | |
2XXU
 
 | | Crystal structure of the GluK2 (GluR6) M770K LBD dimer in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
