4FAN
 
 | |
4FB1
 
 | |
4FAV
 
 | |
4FA9
 
 | |
4FA5
 
 | |
4FA1
 
 | |
4FA4
 
 | |
1ML2
 
 | | Crystal Structure of a Mutant Variant of Cytochrome c Peroxidase with Zn(II)-(20-oxo-Protoporphyrin IX) | | Descriptor: | 20-OXO-PROTOPORPHYRIN IX CONTAINING ZN(II), Cytochrome c Peroxidase | | Authors: | Bhaskar, B, Immoos, C.E, Shimizu, H, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Novel Heme and Peroxide-Dependent Tryptophan-Tyrosine Cross-Link in a Mutant of Cytochrome c Peroxidase
J.Mol.Biol., 328, 2003
|
|
4WX2
 
 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with two F6F molecules in the alpha-site and one F6F molecule in the beta-site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
5JHX
 
 | | Crystal Structure of Fungal MagKatG2 at pH 3.0 | | Descriptor: | CITRATE ANION, Catalase-peroxidase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gasselhuber, B, Obinger, C, Carpena, X. | | Deposit date: | 2016-04-21 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interaction with the Redox Cofactor MYW and Functional Role of a Mobile Arginine in Eukaryotic Catalase-Peroxidase.
Biochemistry, 55, 2016
|
|
8ZCJ
 
 | | Cryo-EM structure of the pasireotide-bound SSTR5-Gi complex | | Descriptor: | 004-DTR-LYS-TYR-PHA-HYP, Beta-2 adrenergic receptor,Somatostatin receptor type 5,lgbit (fusion protein), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y.G, Meng, X.Y, Yang, X.R, Ling, S.L, Shi, P, Tian, C.L, Yang, F. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insights into somatostatin receptor 5 bound with cyclic peptides.
Acta Pharmacol.Sin., 2024
|
|
2DQM
 
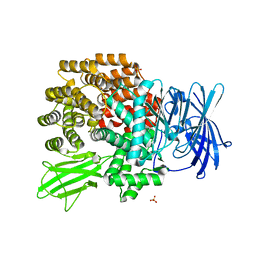 | | Crystal Structure of Aminopeptidase N complexed with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, SULFATE ION, ... | | Authors: | Onohara, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-29 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
2DQ6
 
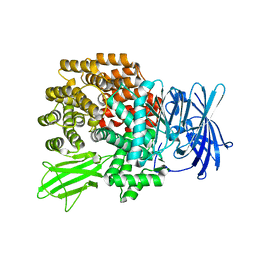 | | Crystal Structure of Aminopeptidase N from Escherichia coli | | Descriptor: | Aminopeptidase N, SULFATE ION, ZINC ION | | Authors: | Nakajima, Y, Onohara, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
8JZ4
 
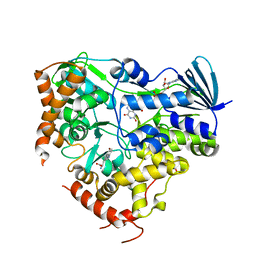 | | Crystal structure of AetF in complex with FAD and 5-bromo-L-tryptophan | | Descriptor: | 5-bromo-L-tryptophan, AetF, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
8JZ3
 
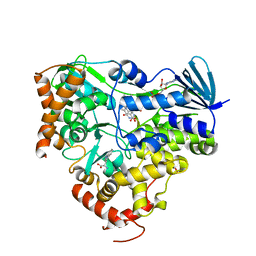 | | Crystal structure of AetF in complex with FAD and L-tryptophan | | Descriptor: | AetF, FLAVIN-ADENINE DINUCLEOTIDE, TRYPTOPHAN | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
2FNM
 
 | | Activation of human carbonic anhdyrase II by exogenous proton donors | | Descriptor: | ZINC ION, carbonic anhydrase 2 | | Authors: | Bhatt, D, Fisher, S.Z, Tu, C, McKenna, R, Silverman, D.N. | | Deposit date: | 2006-01-11 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Location of binding sites in small molecule rescue of human carbonic anhydrase II.
Biophys.J., 92, 2007
|
|
2FNK
 
 | | Activation of Human Carbonic Anhydrase II by exogenous proton donors | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Bhatt, D, Fisher, S.Z, Tu, C, McKenna, R, Silverman, D.N. | | Deposit date: | 2006-01-11 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Location of binding sites in small molecule rescue of human carbonic anhydrase II.
Biophys.J., 92, 2007
|
|
2FNN
 
 | | Activation of human carbonic anhydrase II by exogenous proton donors | | Descriptor: | 4-METHYLIMIDAZOLE, Carbonic Anhydrase 2, ZINC ION | | Authors: | Bhatt, D, Fisher, S.Z, Tu, C, McKenna, R, Silverman, D.N. | | Deposit date: | 2006-01-11 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Location of binding sites in small molecule rescue of human carbonic anhydrase II.
Biophys.J., 92, 2007
|
|
3NUE
 
 | | The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from mycobacterium tuberculosis complexed with tryptophan | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Parker, E.J, Jameson, G.B, Jiao, W, Webby, C.J, Baker, E.N, Baker, H.M. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
J.Biol.Chem., 285, 2010
|
|
2VJQ
 
 | | Formyl-CoA transferase mutant variant W48Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMYL-COENZYME A TRANSFERASE | | Authors: | Toyota, C.G, Berthold, C.L, Gruez, A, Jonsson, S, Lindqvist, Y, Cambillau, C, Richards, N.G.J. | | Deposit date: | 2007-12-11 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential Substrate Specificity and Kinetic Behavior of Escherichia Coli Yfdw and Oxalobacter Formigenes Formyl Coenzyme a Transferase.
J.Bacteriol., 190, 2008
|
|
4A0M
 
 | | CRYSTAL STRUCTURE OF BETAINE ALDEHYDE DEHYDROGENASE FROM SPINACH IN COMPLEX WITH NAD | | Descriptor: | BETAINE ALDEHYDE DEHYDROGENASE, CHLOROPLASTIC, GLYCEROL, ... | | Authors: | Gonzalez-Segura, L, Rudino-Pinera, E, Diaz-Sanchez, A.G, Munoz-Clares, R.A. | | Deposit date: | 2011-09-09 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amino Acid Residues Critical for the Specificity for Betaine Aldehyde of the Plant Aldh10 Isoenzyme Involved in the Synthesis of Glycine Betaine.
Plant Physiol., 158, 2012
|
|
1VYS
 
 | | STRUCTURE OF PENTAERYTHRITOL TETRANITRATE REDUCTASE W102Y MUTANT AND COMPLEXED WITH PICRIC ACID | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-05 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
1VYP
 
 | | Structure of pentaerythritol tetranitrate reductase W102F mutant and complexed with picric acid | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
3RZI
 
 | | The structure of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from mycobacterium tuberculosis cocrystallized and complexed with phenylalanine and tryptophan | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jiao, W, Jameson, G.B, Hutton, R.D, Parker, E.J. | | Deposit date: | 2011-05-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dynamic cross-talk among remote binding sites: the molecular basis for unusual synergistic allostery.
J.Mol.Biol., 415, 2012
|
|
1MKR
 
 | | Crystal Structure of a Mutant Variant of Cytochrome c Peroxidase (Plate like crystals) | | Descriptor: | Cytochrome c Peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Immoos, C.E, Shimizu, H, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Novel Heme and Peroxide-Dependent Tryptophan-Tyrosine Cross-Link in a Mutant of Cytochrome c Peroxidase
J.Mol.Biol., 328, 2003
|
|
