2MOL
 
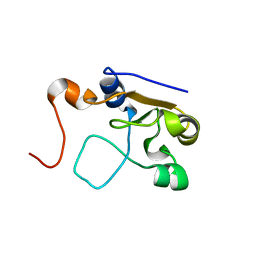 | | 3D NMR structure of the cytoplasmic rhodanese domain of the full-length inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-27 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
7OIN
 
 | | Crystal structure of LSSmScarlet - a genetically encoded red fluorescent protein with a large Stokes shift | | Descriptor: | LSSmScarlet - Genetically Encoded Red Fluorescent Proteins with a Large Stokes Shift, SODIUM ION, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Dorovatovskii, P.V, Subach, O.M, Vlaskina, A.V, Agapova, Y.K, Ivashkina, O.I, Popov, V.O, Subach, F.V. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | LSSmScarlet, dCyRFP2s, dCyOFP2s and CRISPRed2s, Genetically Encoded Red Fluorescent Proteins with a Large Stokes Shift.
Int J Mol Sci, 22, 2021
|
|
1SGE
 
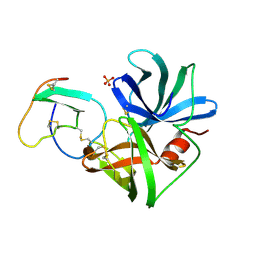 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
1SGQ
 
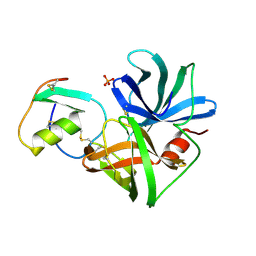 | |
8CWW
 
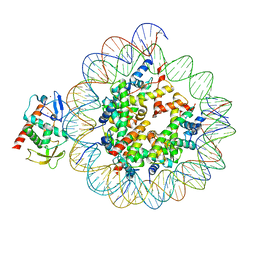 | | Structure of S. cerevisiae Hop1 CBR bound to a nucleosome | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Gu, Y, Ur, S.N, Milano, C.R, Tromer, E.C, Vale-Silva, L.A, Hochwagen, A, Corbett, K.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Chromatin binding by HORMAD proteins regulates meiotic recombination initiation.
Embo J., 43, 2024
|
|
1SAL
 
 | |
2MZE
 
 | | NMR Solution Structure of the PRO Form of Human Matrilysin (proMMP-7) | | Descriptor: | CALCIUM ION, Matrilysin, ZINC ION | | Authors: | Prior, S.H, Fulcher, Y.G, Van Doren, S.R. | | Deposit date: | 2015-02-11 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Charge-Triggered Membrane Insertion of Matrix Metalloproteinase-7, Supporter of Innate Immunity and Tumors.
Structure, 23, 2015
|
|
1AD0
 
 | | FAB FRAGMENT OF ENGINEERED HUMAN MONOCLONAL ANTIBODY A5B7 | | Descriptor: | ANTIBODY A5B7 (HEAVY CHAIN), ANTIBODY A5B7 (LIGHT CHAIN) | | Authors: | Banfield, M.J, Brady, R.L. | | Deposit date: | 1997-02-19 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs.
Proteins, 29, 1997
|
|
8CZE
 
 | | Structure of a Xenopus Nucleosome with Widom 601 DNA | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Gu, Y, Ur, S.N, Milano, C.R, Tromer, E.C, Vale-Silva, L.A, Hochwagen, A, Corbett, K.D. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Chromatin binding by HORMAD proteins regulates meiotic recombination initiation.
Embo J., 43, 2024
|
|
5O5P
 
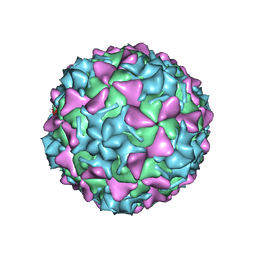 | | Poliovirus type 3 (strain Saukett) stabilized virus-like particle in complex with the pocket factor compound GPP3 | | Descriptor: | 1-[5-[4-(ethoxyiminomethyl)phenoxy]-3-methyl-pentyl]-3-pyridin-4-yl-imidazol-2-one, Capsid proteins, VP4, ... | | Authors: | Bahar, M.W, Kotecha, A, Fry, E.E, Stuart, D.I. | | Deposit date: | 2017-06-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Plant-made polio type 3 stabilized VLPs-a candidate synthetic polio vaccine.
Nat Commun, 8, 2017
|
|
2MPT
 
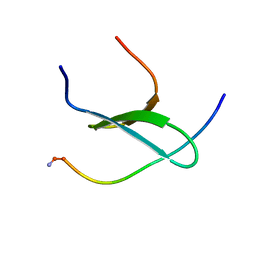 | | WW3 domain of Nedd4L in complex with its HECT domain PY motif | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Escobedo, A, Macias, M.J, Gomes, T, Aragon, E, Martin-Malpartida, P, Ruiz, L. | | Deposit date: | 2014-06-02 | | Release date: | 2014-10-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Activation and Degradation Mechanisms of the E3 Ubiquitin Ligase Nedd4L.
Structure, 22, 2014
|
|
1U7K
 
 | | Structure of a hexameric N-terminal domain from murine leukemia virus capsid | | Descriptor: | Gag polyprotein | | Authors: | Mortuza, G.B, Haire, L.F, Stevens, A, Smerdon, S.J, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution structure of a retroviral capsid hexameric amino-terminal domain.
Nature, 431, 2004
|
|
4QQV
 
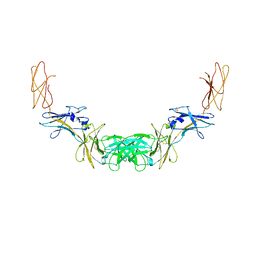 | | Extracellular domains of mouse IL-3 beta receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-3 receptor class 2 subunit beta | | Authors: | Jackson, C.J, Young, I.G, Murphy, J.M, Carr, P.D, Ewens, C.L, Dai, J, Ollis, D.L. | | Deposit date: | 2014-06-30 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of the mouse interleukin-3 beta-receptor: insights into interleukin-3 binding and receptor activation.
Biochem.J., 463, 2014
|
|
1TRL
 
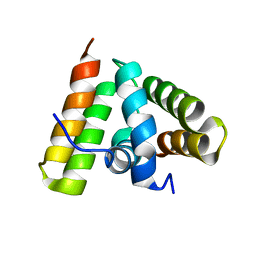 | | NMR SOLUTION STRUCTURE OF THE C-TERMINAL FRAGMENT 255-316 OF THERMOLYSIN: A DIMER FORMED BY SUBUNITS HAVING THE NATIVE STRUCTURE | | Descriptor: | THERMOLYSIN FRAGMENT 255 - 316 | | Authors: | Rico, M, Jimenez, M.A, Gonzalez, C, De Filippis, V, Fontana, A. | | Deposit date: | 1994-09-02 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the C-terminal fragment 255-316 of thermolysin: a dimer formed by subunits having the native structure.
Biochemistry, 33, 1994
|
|
4R39
 
 | |
8AHS
 
 | |
1TRO
 
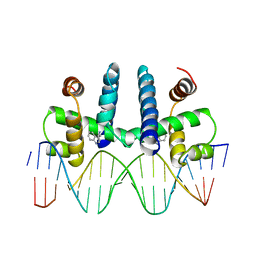 | | CRYSTAL STRUCTURE OF TRP REPRESSOR OPERATOR COMPLEX AT ATOMIC RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*T P*AP*C)-3'), PROTEIN (TRP REPRESSOR), TRYPTOPHAN | | Authors: | Otwinowski, Z, Schevitz, R.W, Zhang, R.-G, Lawson, C.L, Joachimiak, A, Marmorstein, R, Luisi, B.F, Sigler, P.B. | | Deposit date: | 1992-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of trp repressor/operator complex at atomic resolution.
Nature, 335, 1988
|
|
2MUK
 
 | |
4R87
 
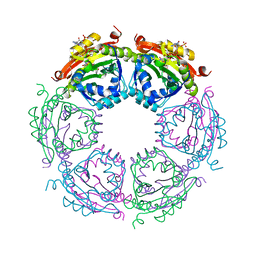 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with CoA and spermine | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, SPERMINE, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-29 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
2N1K
 
 | |
4QTU
 
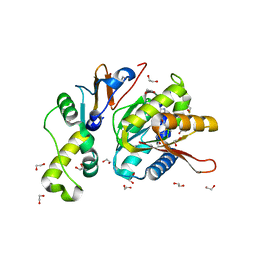 | | Structure of S. cerevisiae Bud23-Trm112 complex involved in formation of m7G1575 on 18S rRNA (SAM bound form) | | Descriptor: | 1,2-ETHANEDIOL, Multifunctional methyltransferase subunit TRM112, Putative methyltransferase BUD23, ... | | Authors: | Letoquart, J, Huvelle, E, Wacheul, L, Bourgeois, G, Zorbas, C, Graille, M, Heurgue-Hamard, V, Lafontaine, D.L.J. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1X9C
 
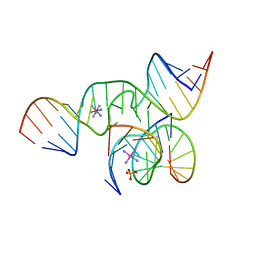 | | An all-RNA Hairpin Ribozyme with mutation U39C | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2004-08-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Conformational Heterogeneity at Position U37 of an All-RNA Hairpin Ribozyme with Implications for Metal Binding and the Catalytic Structure of the S-Turn.
Biochemistry, 44, 2005
|
|
1PL5
 
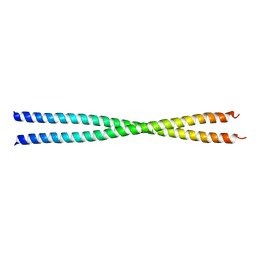 | | Crystal Structure Analysis of the Sir4p C-terminal Coiled Coil | | Descriptor: | Regulatory protein SIR4 | | Authors: | Murphy, G.A, Spedale, E.J, Powell, S.T, Pillus, L, Schultz, S.C, Chen, L. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Sir4 C-terminal coiled coil is required for telomeric and mating type silencing in Saccharomyces cerevisiae.
J.Mol.Biol., 334, 2003
|
|
4YLU
 
 | | X-ray structure of MERS-CoV nsp5 protease bound with a non-covalent inhibitor | | Descriptor: | ACETATE ION, N-{4-[(1H-benzotriazol-1-ylacetyl)(thiophen-3-ylmethyl)amino]phenyl}propanamide, ORF1a protein | | Authors: | Tomar, S, Mesecar, A.D. | | Deposit date: | 2015-03-05 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro): IMPLICATIONS FOR nsp5 REGULATION AND THE DEVELOPMENT OF ANTIVIRALS.
J.Biol.Chem., 290, 2015
|
|
8E2L
 
 | | Structure of Lates calcarifer Twinkle helicase with ATP and DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Gao, Y, Li, Z. | | Deposit date: | 2022-08-15 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural and dynamic basis of DNA capture and translocation by mitochondrial Twinkle helicase.
Nucleic Acids Res., 50, 2022
|
|
