5E7W
 
 | | X-ray Structure of Human Recombinant 2Zn insulin at 0.92 Angstrom | | Descriptor: | ACETATE ION, Insulin, N-PROPANOL, ... | | Authors: | Lisgarten, D.R, Naylor, C.E, Palmer, R.A, Lobley, C.M.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.9519 Å) | | Cite: | Ultra-high resolution X-ray structures of two forms of human recombinant insulin at 100 K.
Chem Cent J, 11, 2017
|
|
5E85
 
 | | isolated SBD of BiP | | Descriptor: | 78 kDa glucose-regulated protein, SULFATE ION | | Authors: | Liu, Q, Yang, J, Nune, M, Zong, Y, Zhou, L. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP.
Structure, 23, 2015
|
|
5E95
 
 | | Crystal Structure of Mb(NS1)/H-Ras Complex | | Descriptor: | GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eguchi, R.R, Sha, F, Gupta, A, Koide, A, Koide, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Inhibition of RAS function through targeting an allosteric regulatory site.
Nat. Chem. Biol., 13, 2017
|
|
5EAC
 
 | | Saccharomyces cerevisiae CYP51 complexed with the plant pathogen inhibitor R-tebuconazole | | Descriptor: | (3~{R})-1-(4-chlorophenyl)-4,4-dimethyl-3-(1,2,4-triazol-1-ylmethyl)pentan-3-ol, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tyndall, J.D.A, Sabherwal, M, Sagatova, A.A, Keniya, M.V, Wilson, R.K, Woods, M.V, Monk, B.C. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and Functional Elucidation of Yeast Lanosterol 14 alpha-Demethylase in Complex with Agrochemical Antifungals.
PLoS ONE, 11, 2016
|
|
7C36
 
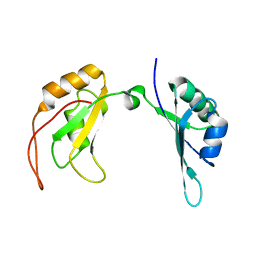 | | c-Myc DNA binding protein structure | | Descriptor: | RNA-binding motif, single-stranded-interacting protein 1 | | Authors: | Aggarwal, P, Bhavesh, N.S. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Hinge like domain motion facilitates human RBMS1 protein binding to proto-oncogene c-myc promoter.
Nucleic Acids Res., 49, 2021
|
|
6DH3
 
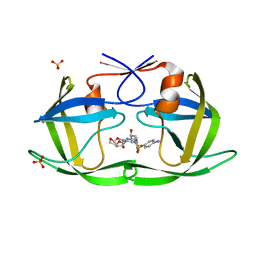 | | Crystal structure of HIV-1 Protease NL4-3 V82I Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.
ACS Infect Dis, 5, 2019
|
|
7CJW
 
 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a crowded environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
3MSJ
 
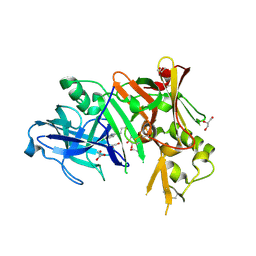 | | Structure of bace (beta secretase) in complex with inhibitor | | Descriptor: | 3-(2-amino-5-chloro-1H-benzimidazol-1-yl)propan-1-ol, BETA-SECRETASE 1, GLYCEROL | | Authors: | Madden, J, Kramer, J, Smith, M.A, Barker, J, Godemann, R. | | Deposit date: | 2010-04-29 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4C9M
 
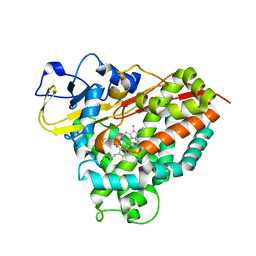 | |
3M3U
 
 | |
4BY0
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-3,3'- difluoro-(1,1'-biphenyl)-4-carboxamide | | Descriptor: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-3,3'-difluoro-(1,1'-biphenyl)-4-carboxamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Choi, J.Y, Calvet, C.M, Vierira, D.F, Gunatilleke, S.S, Cameron, M.D, McKerrow, J.H, Podust, L.M, Roush, W.R. | | Deposit date: | 2013-07-16 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | R-Configuration of 4-Aminopyridyl-Based Inhibitors of Cyp51 Confers Superior Efficacy Against Trypanosoma Cruzi
Acs Med.Chem.Lett., 5, 2014
|
|
4C0C
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(2,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | Descriptor: | 4-[4-[2,4-bis(fluoranyl)phenyl]piperazin-1-yl]-2-fluoranyl-N-[(2R)-3-(1H-indol-3-yl)-1-oxidanylidene-1-(pyridin-4-ylamino)propan-2-yl]benzamide, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Calvet, C.M, Vieira, D.F, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2013-08-01 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | 4-Aminopyridyl-Based Cyp51 Inhibitors as Anti-Trypanosoma Cruzi Drug Leads with Improved Pharmacokinetic Profile and in Vivo Potency.
J.Med.Chem., 57, 2014
|
|
4C9K
 
 | |
7DIH
 
 | |
3MDT
 
 | | Voriconazole complex of Cytochrome P450 46A1 | | Descriptor: | Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
4C9N
 
 | |
6DWS
 
 | | Crystal structure of human GRP78 in complex with (2R,3R,4S,5R)-2-(6-amino-8-((2-chlorobenzyl)amino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol | | Descriptor: | 8-{[(2-chlorophenyl)methyl]amino}adenosine, Endoplasmic reticulum chaperone BiP, MAGNESIUM ION | | Authors: | Chen, Y, Antoshchenko, T, Smil, D, Zepeda, C, Huang, Y, Park, H.W. | | Deposit date: | 2018-06-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human GRP78 in complex with (2R,3R,4S,5R)-2-(6-amino-8-((2-chlorobenzyl)amino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol
To Be Published
|
|
3MF4
 
 | | Crystal structure of putative two-component system response regulator/ggdef domain protein | | Descriptor: | MAGNESIUM ION, Two-component system response regulator/GGDEF domain protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-01 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative two-component system response regulator/ggdef domain protein
To be Published
|
|
3MJH
 
 | | Crystal Structure of Human Rab5A in complex with the C2H2 Zinc Finger of EEA1 | | Descriptor: | Early endosome antigen 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mishra, A.K, Eathiraj, S, Lambright, D.G. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of Early Endosomal Autoantigen 1 (EEA1).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4C3V
 
 | | Extensive counter-ion interactions with subtilisin in aqueous medium, no Cs soak | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN CARLSBERG, ... | | Authors: | Cianci, M, Negroni, J, Helliwell, J.R, Halling, P.J. | | Deposit date: | 2013-08-27 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Extensive Counter-Ion Interactions Seen at the Surface of Subtilisin in an Aqueous Medium
Rsc Adv, 2014
|
|
3M0U
 
 | |
3MOU
 
 | |
4C9O
 
 | |
4CBO
 
 | |
7DD0
 
 | |
