7OFS
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 4-(2-hydroxyethyl)phenol | | Descriptor: | 4-(2-hydroxyethyl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Ewert, W, Sprenger, J, Koua, F, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Wolf, M, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFT
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with p-hydroxybenzaldehyde | | Descriptor: | CHLORIDE ION, P-HYDROXYBENZALDEHYDE, POTASSIUM ION, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Ewert, W, Sprenger, J, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Wolf, M, Yefanov, O, Gelisio, L, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
6YPW
 
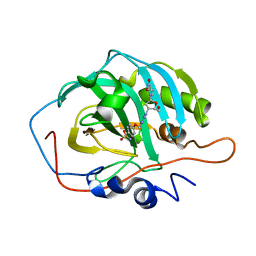 | | Crystal structure for the complex of human carbonic anhydrase II and 4-((1-(2-(hydroxymethyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-1H-1,2,3-triazol-4-yl)methoxy)benzenesulfonamide | | Descriptor: | 4-[[1-[(2~{S},3~{S},5~{R})-2-(hydroxymethyl)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]oxolan-3-yl]-1,2,3-triazol-4-yl]methoxy]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2020-04-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mechanisms of the Antiproliferative and Antitumor Activity of Novel Telomerase-Carbonic Anhydrase Dual-Hybrid Inhibitors.
J.Med.Chem., 64, 2021
|
|
1SNB
 
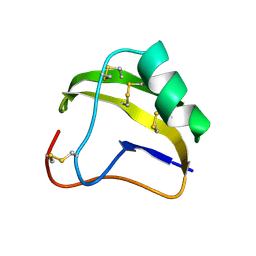 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M8 | | Descriptor: | NEUROTOXIN BMK M8 | | Authors: | Wang, D.C, Zeng, Z.H, Li, H.M. | | Deposit date: | 1997-03-12 | | Release date: | 1997-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an acidic neurotoxin from scorpion Buthus martensii Karsch at 1.85 A resolution.
J.Mol.Biol., 261, 1996
|
|
4O4S
 
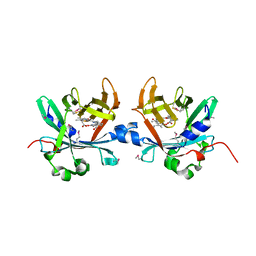 | | Crystal structure of phycobiliprotein lyase CpcT complexed with phycocyanobilin (PCB) | | Descriptor: | PHYCOCYANOBILIN, Phycocyanobilin lyase CpcT | | Authors: | Zhou, W, Ding, W.-L, Zeng, X.-l, Dong, L.-L, Zhao, B, Zhou, M, Scheer, H, Zhao, K.-H, Yang, X. | | Deposit date: | 2013-12-19 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of the Phycobiliprotein Lyase CpcT.
J.Biol.Chem., 289, 2014
|
|
4O4O
 
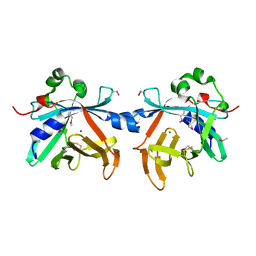 | | Crystal structure of phycobiliprotein lyase CpcT | | Descriptor: | MAGNESIUM ION, Phycocyanobilin lyase CpcT | | Authors: | Zhou, W, Ding, W.-L, Zeng, X.-l, Dong, L.-L, Zhao, B, Zhou, M, Scheer, H, Zhao, K.-H, Yang, X. | | Deposit date: | 2013-12-19 | | Release date: | 2014-08-06 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Mechanism of the Phycobiliprotein Lyase CpcT.
J.Biol.Chem., 289, 2014
|
|
6GOT
 
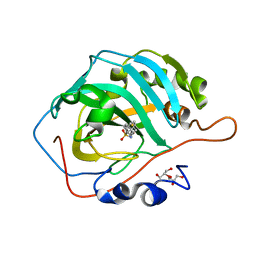 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor 4-(phenethylthio)benzenesulfonamide | | Descriptor: | 4-(2-phenylethylsulfanyl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Angeli, A. | | Deposit date: | 2018-06-04 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Synthesis of different thio-scaffolds bearing sulfonamide with subnanomolar carbonic anhydrase II and IX inhibitory properties and X-ray investigations for their inhibitory mechanism.
Bioorg. Chem., 81, 2018
|
|
1SN4
 
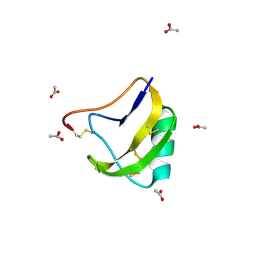 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1MHL
 
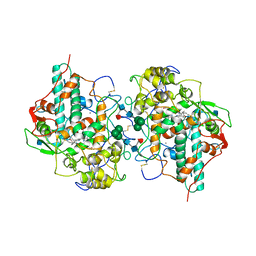 | | CRYSTAL STRUCTURE OF HUMAN MYELOPEROXIDASE ISOFORM C CRYSTALLIZED IN SPACE GROUP P2(1) AT PH 5.5 AND 20 DEG C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fenna, R.E, Zeng, J, Davey, C. | | Deposit date: | 1995-06-09 | | Release date: | 1996-01-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the green heme in myeloperoxidase.
Arch.Biochem.Biophys., 316, 1995
|
|
1SN1
 
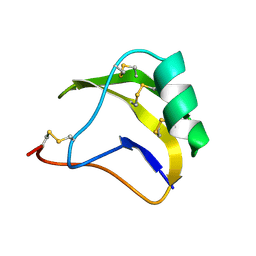 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
6GDC
 
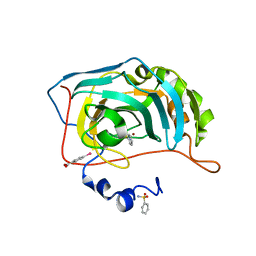 | | Human Carbonic Anhydrase II in complex with Benzenesulfonamide | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, MERCURY (II) ION, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.079 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
7KZE
 
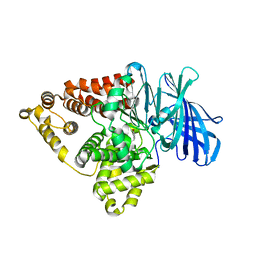 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | Descriptor: | 1-benzyl-4-methoxybenzene, Leukotriene A-4 hydrolase, TRIETHYLENE GLYCOL, ... | | Authors: | Lee, K.H, Shim, Y, Paige, M, Noble, S.M. | | Deposit date: | 2020-12-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
1SM1
 
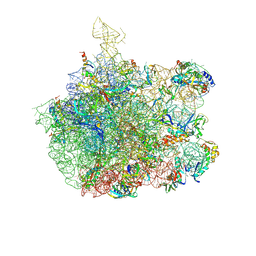 | | COMPLEX OF THE LARGE RIBOSOMAL SUBUNIT FROM DEINOCOCCUS RADIODURANS WITH QUINUPRISTIN AND DALFOPRISTIN | | Descriptor: | 23S RIBOSOMAL RNA, 5-(2-DIETHYLAMINO-ETHANESULFONYL)-21-HYDROXY-10-ISOPROPYL-11,19-DIMETHYL-9,26-DIOXA-3,15,28-TRIAZA-TRICYCLO[23.2.1.00,255]OCTACOSA-1(27),12,17,19,25(28)-PENTAENE-2,8,14,23-TETRAONE, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Harms, J.M, Schluenzen, F, Fucini, P, Bartels, H, Yonath, A. | | Deposit date: | 2004-03-08 | | Release date: | 2004-08-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Alterations at the Peptidyl Transferase Centre of the Ribosome Induced by the Synergistic Action of the Streptogramins Dalfopristin and Quinupristin.
Bmc Biol., 2, 2004
|
|
8ZR5
 
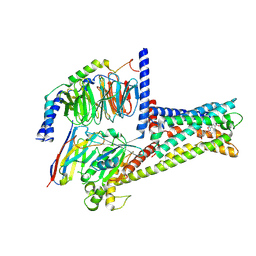 | | Cryo-EM Structure of GPR119-Gs-Firuglipel complex | | Descriptor: | 4-[5-[(1~{R})-1-(4-cyclopropylcarbonylphenoxy)propyl]-1,2,4-oxadiazol-3-yl]-2-fluoranyl-~{N}-[(2~{R})-1-oxidanylpropan-2-yl]benzamide, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, Qiu, C, Du, Y. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of GPR119-Gs complex
To Be Published
|
|
8ZRK
 
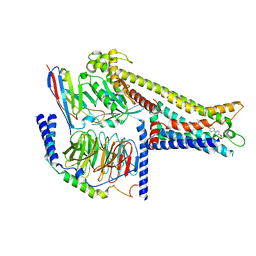 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist GSK-1292263 | | Descriptor: | 5-[4-[[6-(4-methylsulfonylphenyl)pyridin-3-yl]oxymethyl]piperidin-1-yl]-3-propan-2-yl-1,2,4-oxadiazole, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wong, T.S, Xiong, T.T, Zeng, Z.C, Gan, S.Y, Qiu, C, Du, Y. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM structure of GPR119-Gs complex
To be published
|
|
6TPE
 
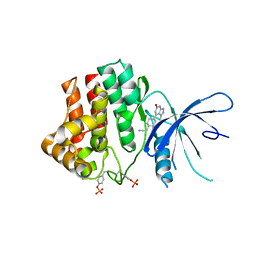 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | Descriptor: | 2-[4-(3-methyl-6-oxidanylidene-1,7-dihydropyrazolo[3,4-b]pyridin-4-yl)cyclohexyl]ethanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Hansen, B.B, Jepsen, T.H, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Molck, C, Rai, S, Nasipireddy, V.R, Jestel, A, Lammens, A, Ritzen, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
4HLA
 
 | | Crystal structure of wild type HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2012-10-16 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
6TPF
 
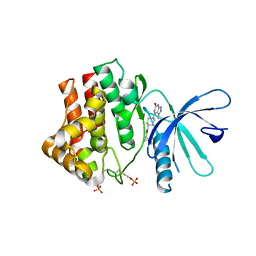 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | Descriptor: | (1~{S})-2,2-bis(fluoranyl)-~{N}-[4-(3-methyl-6-oxidanylidene-2,7-dihydropyrazolo[3,4-b]pyridin-4-yl)cyclohexyl]cyclopropane-1-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Hansen, B.B, Jepsen, T.H, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Molck, C, Rai, S, Nasipireddy, V.R, Griessner, A, Ritzen, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
5MCU
 
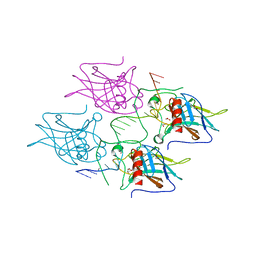 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LHG2) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
5MCV
 
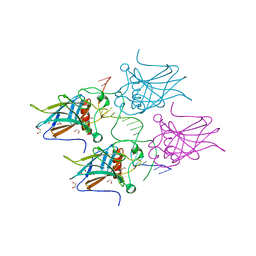 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LWC1) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cellular tumor antigen p53, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
5MI8
 
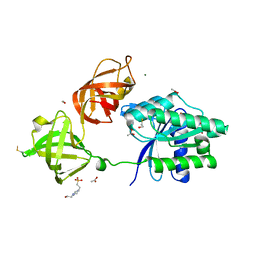 | | Structure of the phosphomimetic mutant of EF-Tu T383E | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Talavera, A, Hendrix, J, Versees, W, De Gieter, S, Castro-Roa, D, Jurenas, D, Van Nerom, K, Vandenberk, N, Barth, A, De Greve, H, Hofkens, J, Zenkin, N, Loris, R, Garcia-Pino, A. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors.
Sci Adv, 4, 2018
|
|
4I7J
 
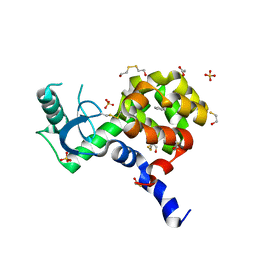 | | T4 Lysozyme L99A/M102H with benzene bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BENZENE, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
4I8W
 
 | | Crystal structure of wild type HIV-1 protease in complex with non-peptidic inhibitor, GRL007 | | Descriptor: | 4-{[(2R,3S)-3-({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yloxy]carbonyl}amino)-2-hydroxy-4-phenylbutyl](2-methylpropyl)sulfamoyl}benzoic acid, Protease | | Authors: | Yedidi, R.S, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2012-12-04 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
5MCW
 
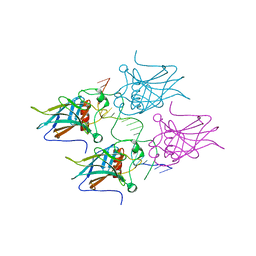 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LWC2) | | Descriptor: | Cellular tumor antigen p53, DNA, FORMYL GROUP, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
5MCT
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LHG1) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
