6OVK
 
 | |
7UDX
 
 | | Designed pentameric proton channel QLQL | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed pentameric proton channel QLQL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7Z76
 
 | | Crystal structure of compound 10 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{R})-2-[(2~{R})-1-[4-(4-bromanyl-7-cyclopentyl-5-oxidanylidene-benzimidazolo[1,2-a]quinazolin-9-yl)piperidin-1-yl]propan-2-yl]oxy-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-1-[(2~{S})-2-[[1-(dimethylamino)cyclopropyl]carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo.
Nat Commun, 13, 2022
|
|
7Z78
 
 | | Crystal structure of compound 4 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | 4-bromanyl-7-cyclopentyl-9-piperidin-4-yl-benzimidazolo[1,2-a]quinazolin-5-one, Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo.
Nat Commun, 13, 2022
|
|
1KDF
 
 | | NORTH-ATLANTIC OCEAN POUT ANTIFREEZE PROTEIN TYPE III ISOFORM HPLC12 MUTANT, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Sonnichsen, F.D, Deluca, C.I, Davies, P.L, Sykes, B.D. | | Deposit date: | 1996-07-08 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of type III antifreeze protein: hydrophobic groups may be involved in the energetics of the protein-ice interaction.
Structure, 4, 1996
|
|
1KVU
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
7UFV
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-6766 | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chlorophenyl)-3-oxopropyl]-3-(2-fluorophenyl)-1H-pyrazole-4-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Li, A, Li, Y, Dong, A, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nanomolar DCAF1 Small Molecule Ligands.
J.Med.Chem., 66, 2023
|
|
8BFU
 
 | | Crystal structure of the apo p110alpha catalytic subunit from homo sapiens | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gong, G.Q, Bellini, D, Vanhaesebroeck, B, Williams, R.L. | | Deposit date: | 2022-10-26 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
6OWK
 
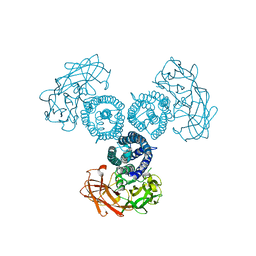 | |
3H5U
 
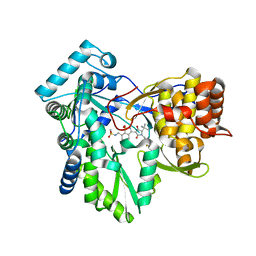 | | Hepatitis C virus polymerase NS5B with saccharin inhibitor 1 | | Descriptor: | N-({3-[(5S)-5-tert-butyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-1,2-benzisothiazol-7-yl}methyl)methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 4: structure-based design, synthesis, and biological evaluation of benzo[d]isothiazole-1,1-dioxides
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3H7T
 
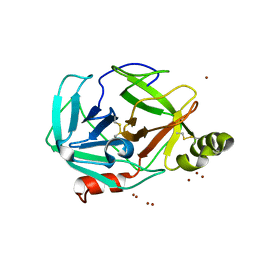 | |
6OVM
 
 | |
3GWE
 
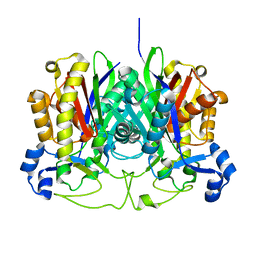 | |
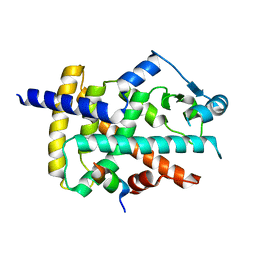
8BF1
 
 | |
7UWG
 
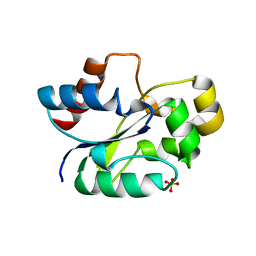 | | The crystal structure of the TIR domain-containing protein from Acinetobacter baumannii (AbTir) | | Descriptor: | HEXAETHYLENE GLYCOL, Molecular chaperone Tir, SULFATE ION | | Authors: | Manik, M.K, Nanson, J.D, Ve, T, Kobe, B. | | Deposit date: | 2022-05-03 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXR
 
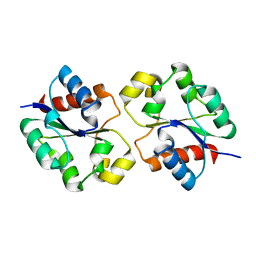 | | Crystal structure of the BtTir TIR domain | | Descriptor: | TIR domain protein | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Vasquez, E, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
6QTZ
 
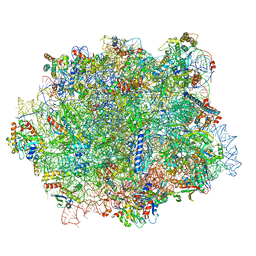 | |
8BFF
 
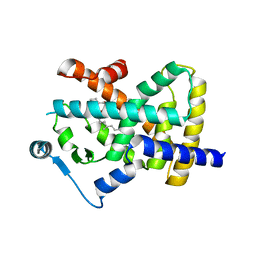 | | Human PPARgamma in complex with MINCH bound to the AF-2 sub-pocket | | Descriptor: | (1~{S},2~{R})-2-[(4~{R})-4-methylheptoxy]carbonylcyclohexane-1-carboxylic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Useini, A, Straeter, N. | | Deposit date: | 2022-10-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the activation of PPAR gamma by the plasticizer metabolites MEHP and MINCH.
Environ Int, 173, 2023
|
|
7UXS
 
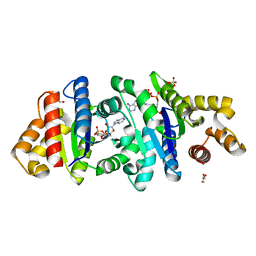 | | Crystal structure of the BcThsA SLOG domain in complex with 3'cADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, BcThsA, GLYCEROL, ... | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
8BF2
 
 | |
3H6A
 
 | |
3H7O
 
 | |
7UXT
 
 | | Crystal structure of ligand-free SeThsA | | Descriptor: | GLYCEROL, TRIETHYLENE GLYCOL, USG protein | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Nanson, J.D, Kobe, B, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
8A8R
 
 | | Crystal structure of TEAD4 in complex with YAP peptide | | Descriptor: | Isoform 7 of Transcriptional coactivator YAP1, MYRISTIC ACID, Transcriptional enhancer factor TEF-3 | | Authors: | Scheufler, C, Kallen, J. | | Deposit date: | 2022-06-23 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | N-terminal beta-strand in YAP is critical for stronger binding to scalloped relative to TEAD transcription factor.
Protein Sci., 32, 2023
|
|
6QO7
 
 | |
