4AXF
 
 | | InsP5 2-K in complex with Ins(3,4,5,6)P4 plus AMPPNP | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, Myo inositol 3,4,5,6 tetrakisphosphate, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
4B7X
 
 | | Crystal structure of hypothetical protein PA1648 from Pseudomonas aeruginosa. | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROBABLE OXIDOREDUCTASE | | Authors: | Alphey, M.S, McMahon, S.A, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4AVY
 
 | | The AEROPATH project and Pseudomonas aeruginosa high-throughput crystallographic studies for assessment of potential targets in early stage drug discovery. | | Descriptor: | PROBABLE SHORT-CHAIN DEHYDROGENASE | | Authors: | Moynie, L, McMahon, S.A, Alphey, M.S, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-05-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4D44
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-ethyl- 4-fluoro-2-((2-fluoropyridin-3-yl)oxy)phenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5-ethyl-4-fluoro-2-[(2-fluoropyridin-3-yl)oxy]phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
6Y4U
 
 | | Crystal structure of p38 in complex with SR65 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-oxidanylidene-1-(pentan-3-ylamino)butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
7A0O
 
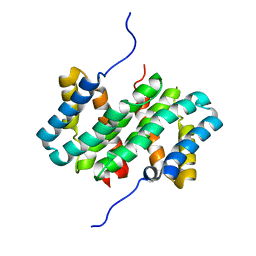 | | NMR structure of flagelliform spidroin (FlagSp) N-terminal domain from Trichonephila clavipes at pH 5.5 | | Descriptor: | Flagelliform spidroin variant 1 | | Authors: | Sarr, M, Kitoka, K, Walsh-White, K.-A, Kaldmae, M, Landreh, M, Rising, A, Johansson, J, Jaudzems, K, Kronqvist, N. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence.
J.Biol.Chem., 298, 2022
|
|
7A0I
 
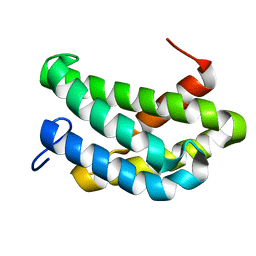 | | NMR structure of flagelliform spidroin (FlagSp) N-terminal domain from Trichonephila clavipes at pH 7.2 | | Descriptor: | Flagelliform spidroin variant 1 | | Authors: | Sarr, M, Kitoka, K, Walsh-White, K.-A, Kaldmae, M, Landreh, M, Rising, A, Johansson, J, Jaudzems, K, Kronqvist, N. | | Deposit date: | 2020-08-09 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence.
J.Biol.Chem., 298, 2022
|
|
6EL6
 
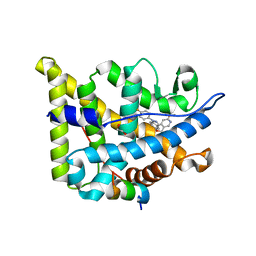 | | Glucocorticoid Receptor in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, Glucocorticoid receptor, Nuclear receptor coactivator 2, ... | | Authors: | Edman, K, Wissler, L. | | Deposit date: | 2017-09-28 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Novel Oral Glucocorticoid Receptor Modulator (AZD9567) with Improved Side Effect Profile.
J. Med. Chem., 61, 2018
|
|
6Y4T
 
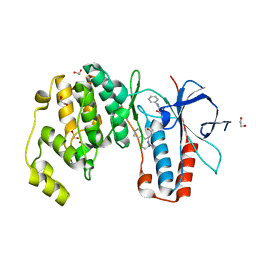 | | Crystal structure of p38 in complex with SR63. | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-1-[[(2~{S})-butan-2-yl]amino]-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
4PNU
 
 | | E. coli sliding clamp in complex with (R)-6-bromo-9-(2-((R)-1-carboxy-2-phenylethylamino)-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-6-bromo-9-(2-{[(1R)-1-carboxy-2-phenylethyl]amino}-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
7ASD
 
 | | Structure of native royal jelly filaments | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mattei, S, Ban, A, Picenoni, A, Leibundgut, M, Glockshuber, R, Boehringer, D. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of native glycolipoprotein filaments in honeybee royal jelly.
Nat Commun, 11, 2020
|
|
4PQH
 
 | | Crystal structure of glutathione transferase lambda1 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, SODIUM ION, glutathione transferase lambda1 | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
6Y4V
 
 | | Crystal structure of p38 in complex with SR68 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(3-methylbutylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
2MJT
 
 | | Anoplin R5F T8W in DPC micelles | | Descriptor: | Anoplin, UNKNOWN ATOM OR ION, dodecyl 2-(trimethylammonio)ethyl phosphate | | Authors: | Uggerhoej, L, Poulsen, T.J, Wimmer, R. | | Deposit date: | 2014-01-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Alpha-Helical Antimicrobial Peptides: Do's and Don'ts.
Chembiochem, 16, 2015
|
|
4PY1
 
 | | Crystal structure of Tyk2 in complex with compound 15, 6-((2,5-dimethoxyphenyl)thio)-3-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine | | Descriptor: | 6-[(2,5-dimethoxyphenyl)sulfanyl]-3-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazine, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Kinase domain inhibition of leucine rich repeat kinase 2 (LRRK2) using a [1,2,4]triazolo[4,3-b]pyridazine scaffold.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6Y6V
 
 | | p38a bound with MCP-81 | | Descriptor: | 5-azanyl-~{N}-[[4-[[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Schroeder, M, Roehm, S, Knapp, S, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
4PZ1
 
 | | Crystal structure of a sHIP (UniProt Id: Q99XU0) mutant from Streptococcus pyogenes | | Descriptor: | CALCIUM ION, CHLORIDE ION, sHIP | | Authors: | Wisniewska, M, Happonen, L, Frick, I.-M, Bjorck, L, Streicher, W, Malmstrom, J, Wikstrom, M. | | Deposit date: | 2014-03-28 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Functional and structural properties of a novel protein and virulence factor (Protein sHIP) in Streptococcus pyogenes.
J.Biol.Chem., 289, 2014
|
|
4DDO
 
 | |
4DH6
 
 | | Structure of Bace-1 (Beta-Secretase) in Complex with (2R)-N-((2S,3R)-1-(benzo[d][1,3]dioxol-5-yl)-3-hydroxy-4-((S)-6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridine]-4'-ylamino)butan-2-yl)-2-methoxypropanamide | | Descriptor: | (2R)-N-[(2S,3R)-1-(1,3-benzodioxol-5-yl)-4-{[(4'S)-6'-(2,2-dimethylpropyl)-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl]amino}-3-hydroxybutan-2-yl]-2-methoxypropanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2012-01-27 | | Release date: | 2012-04-18 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and preparation of a potent series of hydroxyethylamine containing beta-secretase inhibitors that demonstrate robust reduction of central beta-amyloid.
J.Med.Chem., 55, 2012
|
|
6YK7
 
 | | Crystal structure of p38 in complex with SR43 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(ethylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-05 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6EL7
 
 | | Glucocorticoid Receptor in complex with compound 31 | | Descriptor: | 1,2-ETHANEDIOL, Glucocorticoid receptor, Nuclear receptor coactivator 2, ... | | Authors: | Edman, K, Wissler, L. | | Deposit date: | 2017-09-28 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of a Novel Oral Glucocorticoid Receptor Modulator (AZD9567) with Improved Side Effect Profile.
J. Med. Chem., 61, 2018
|
|
4Q4W
 
 | |
4DDD
 
 | |
4DWS
 
 | |
8FMA
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C11 multimer | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
