5VBQ
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRDT IN COMPLEX WITH BI2536 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain testis-specific protein, ... | | Authors: | EMBER, S.W, ZHU, J.-Y, SCHONBRUNN, E. | | Deposit date: | 2017-03-30 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 2021
|
|
5VBR
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRDT IN COMPLEX WITH Volasertib | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain testis-specific protein, CHLORIDE ION, ... | | Authors: | EMBER, S.W, ZHU, J.-Y, SCHONBRUNN, E. | | Deposit date: | 2017-03-30 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 2021
|
|
1MHO
 
 | | THE 2.0 A STRUCTURE OF HOLO S100B FROM BOVINE BRAIN | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Matsumura, H, Shiba, T, Inoue, T, Harada, S, Yasushi, K.A.I. | | Deposit date: | 1997-09-11 | | Release date: | 1998-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain.
Structure, 6, 1998
|
|
5HPK
 
 | | System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes: NEDD4L and UbV NL.1 | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like, Ubiquitin variant NL.1 | | Authors: | Wu, K.-P, Mukherjee, M, Mercredi, P.Y, Schulman, B.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
1ZMQ
 
 | | Crystal structure of human alpha-defensin-6 | | Descriptor: | CHLORIDE ION, Defensin 6 | | Authors: | Lubkowski, J, Szyk, A, Lu, W. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human {alpha}-defensins HNP4, HD5, and HD6.
Protein Sci., 15, 2006
|
|
5IB3
 
 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR and Copper | | Descriptor: | Beta-2-microglobulin, COPPER (II) ION, GLYCEROL, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
1ZMP
 
 | | Crystal structure of human defensin-5 | | Descriptor: | CHLORIDE ION, Defensin 5, GLYCEROL, ... | | Authors: | Lubkowski, J, Szyk, A, Lu, W. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of human {alpha}-defensins HNP4, HD5, and HD6.
Protein Sci., 15, 2006
|
|
3B3I
 
 | | Citrullination-dependent differential presentation of a self-peptide by HLA-B27 subtypes | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Beltrami, A, Rossmann, M, Fiorillo, M.T, Paladini, F, Sorrentino, R, Saenger, W, Kumar, P, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2007-10-22 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Citrullination-dependent differential presentation of a self-peptide by HLA-B27 subtypes.
J.Biol.Chem., 283, 2008
|
|
4C66
 
 | | Discovery of Epigenetic Regulator I-BET762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the BET Bromodomains | | Descriptor: | 4-(2-chlorophenyl)-2-ethyl-9-methyl-6,8-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Mirguet, O. | | Deposit date: | 2013-09-17 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Epigenetic Regulator I-Bet762: Lead Optimization to Afford a Clinical Candidate Inhibitor of the Bet Bromodomains.
J.Med.Chem., 56, 2013
|
|
5WY5
 
 | | Crystal structure of MAGEG1 and NSE1 complex | | Descriptor: | MAGNESIUM ION, Melanoma-associated antigen G1, Non-structural maintenance of chromosomes element 1 homolog, ... | | Authors: | Yang, M, Gao, J. | | Deposit date: | 2017-01-11 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Mage-Ring Protein Complexes Comprise A Family Of E3 Ubiquitin Ligases.
Mol.Cell, 39, 2010
|
|
1Z95
 
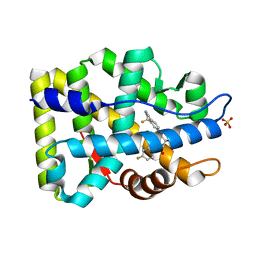 | | Crystal Structure of the Androgen Receptor Ligand-binding Domain W741L Mutant Complex with R-bicalutamide | | Descriptor: | Androgen Receptor, R-BICALUTAMIDE, SULFATE ION | | Authors: | Bohl, C.E, Gao, W, Miller, D.D, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-03-31 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for antagonism and resistance of bicalutamide in prostate cancer.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7LV7
 
 | |
1W01
 
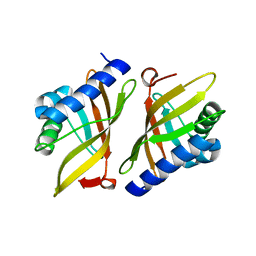 | |
1W00
 
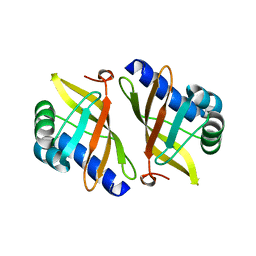 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 2004-05-30 | | Release date: | 2005-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
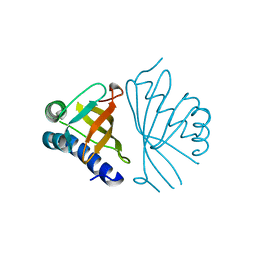
1W02
 
 | |
2PNU
 
 | | Crystal structure of human androgen receptor ligand-binding domain in complex with EM-5744 | | Descriptor: | (5S,8R,9S,10S,13R,14S,17S)-13-{2-[(3,5-DIFLUOROBENZYL)OXY]ETHYL}-17-HYDROXY-10-METHYLHEXADECAHYDRO-3H-CYCLOPENTA[A]PHENANTHREN-3-ONE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Cantin, L, Faucher, F, Couture, J.F, Pereira de Jesus-Tran, K, Legrand, P, Ciobanu, C.L, Singh, S.M, Labrie, F, Breton, R. | | Deposit date: | 2007-04-25 | | Release date: | 2007-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of the Human Androgen Receptor Ligand-binding Domain Complexed with EM5744, a Rationally Designed Steroidal Ligand Bearing a Bulky Chain Directed toward Helix 12.
J.Biol.Chem., 282, 2007
|
|
3H9E
 
 | | Crystal structure of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDS) complex with NAD and phosphate | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W, Cocking, R, Bray, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS.
Biochem.J., 435, 2011
|
|
2Y93
 
 | | Crystal Structure of cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB)from Pandoraea pnomenusa strain B-356. | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-02-11 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
2Y99
 
 | | Crystal Structure of cis-Biphenyl-2,3-dihydrodiol-2,3-dehydrogenase (BphB)from Pandoraea pnomenusa strain B-356 complex with co-enzyme NAD | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-02-12 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
3ZU7
 
 | |
3ZUV
 
 | |
2WA0
 
 | | Crystal structure of the human MAGEA4 | | Descriptor: | MELANOMA-ASSOCIATED ANTIGEN 4 | | Authors: | Roos, A.K, Cooper, C.D.O, Ugochukwu, E, W Yue, W, Berridge, G, Elkins, J.M, Pike, A.C.W, Bray, J, Filippakopoulos, P, Muniz, J, Chaikuad, A, Burgess-Brown, N, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O, Oppermann, U. | | Deposit date: | 2009-01-31 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Two Melanoma-Associated Antigens Suggest Allosteric Regulation of Effector Binding.
Plos One, 11, 2016
|
|
2GW9
 
 | | High-resolution solution structure of the mouse defensin Cryptdin4 | | Descriptor: | Defensin-related cryptdin 4 | | Authors: | Rosengren, K.J, Craik, D.J, Vogel, H.J, Daly, N.L, Ouellette, A.J. | | Deposit date: | 2006-05-04 | | Release date: | 2006-07-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the conserved salt bridge in mammalian paneth cell alpha-defensins: solution structures of mouse CRYPTDIN-4 and (E15D)-CRYPTDIN-4.
J.Biol.Chem., 281, 2006
|
|
2GWP
 
 | | High-resolution solution structure of the salt-bridge defficient mouse defensin (E15D)-Cryptdin4 | | Descriptor: | Defensin-related cryptdin 4 | | Authors: | Rosengren, K.J, Craik, D.J, Vogel, H.J, Daly, N.L, Ouellette, A.J. | | Deposit date: | 2006-05-05 | | Release date: | 2006-07-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the conserved salt bridge in mammalian paneth cell alpha-defensins: solution structures of mouse CRYPTDIN-4 and (E15D)-CRYPTDIN-4.
J.Biol.Chem., 281, 2006
|
|
8KHW
 
 | |
