7MHW
 
 | | Crystal structure of the protease inhibitor U-Omp19 from Brucella abortus fused to Maltose-binding protein | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Outer membrane lipoprotein omp19, SULFATE ION | | Authors: | Darriba, M.L, Klinke, S, Otero, L.H, Cerutti, M.L, Cassataro, J, Pasquevich, K.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A disordered region retains the full protease inhibitor activity and the capacity to induce CD8 + T cells in vivo of the oral vaccine adjuvant U-Omp19.
Comput Struct Biotechnol J, 20, 2022
|
|
5CSY
 
 | | Disproportionating enzyme 1 from Arabidopsis - acarbose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5U16
 
 | | Structure of human MR1-2-OH-1-NA in complex with human MAIT A-F7 TCR | | Descriptor: | 2-hydroxynaphthalene-1-carbaldehyde, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Keller, A.N, Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2016-11-27 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells.
Nat. Immunol., 18, 2017
|
|
7P9C
 
 | | Escherichia coli type II L-asparaginase | | Descriptor: | L-asparaginase 2 | | Authors: | Maggi, M, Scotti, C. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Revealing Escherichia coli type II L-asparaginase active site flexible loop in its open, ligand-free conformation.
Sci Rep, 11, 2021
|
|
7MHD
 
 | | Thioesterase Domain of Human Fatty Acid Synthase (FASN-TE) binding a competitive inhibitor SBP-7635 | | Descriptor: | Fatty acid synthase, N,N-diethyl-4-{2-[(2-fluorophenyl)methyl]-1,3-thiazol-4-yl}benzene-1-sulfonamide | | Authors: | Aleshin, A.E, Lambert, L, Liddington, R.C, Cosford, N. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Thioesterase Domain of Human Fatty Acid Synthase (FASN-TE) binding a competitive inhibitor SBP-7635
To Be Published
|
|
5KAU
 
 | | The structure of SAV2435 bound to RHODAMINE 6G | | Descriptor: | GLYCEROL, RHODAMINE 6G, SA2223 protein | | Authors: | Moreno, A, Wade, H. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Solution Binding and Structural Analyses Reveal Potential Multidrug Resistance Functions for SAV2435 and CTR107 and Other GyrI-like Proteins.
Biochemistry, 55, 2016
|
|
8B1S
 
 | | co-crystal of SUDV VP40 with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, Matrix protein VP40 | | Authors: | Werner, A.-D, Krapoth, N, Norris, M.J, Heine, A, Klebe, G, Ollmann Saphire, E, Becker, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-27 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a Crystallographic Screening to Identify Sudan Virus VP40 Ligands.
Acs Omega, 9, 2024
|
|
5CU6
 
 | | Crystal Structure of CK2alpha | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
8ARX
 
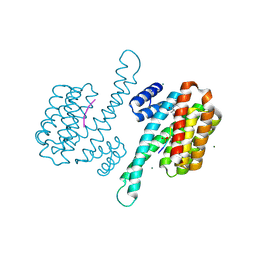 | | Small molecular stabilizer for ERalpha and 14-3-3sigma (1074378) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[1-(4-chloranylphenoxy)cyclopropyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AK4
 
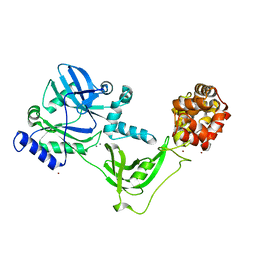 | | Structure of the C-terminally truncated NAD+-dependent DNA ligase from the poly-extremophile Deinococcus radiodurans | | Descriptor: | DNA ligase, MANGANESE (II) ION, ZINC ION | | Authors: | Fernandes, A, Williamson, A.K, Matias, P.M, Moe, E. | | Deposit date: | 2022-07-29 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structure/function studies of the NAD + -dependent DNA ligase from the poly-extremophile Deinococcus radiodurans reveal importance of the BRCT domain for DNA binding.
Extremophiles, 27, 2023
|
|
6LG4
 
 | |
7OWC
 
 | |
5U2M
 
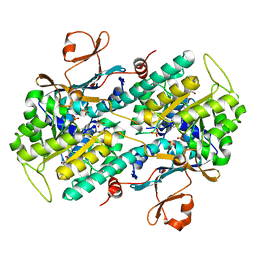 | | Crystal structure of human NAMPT with A-1293201 | | Descriptor: | N-[4-({[(3S)-oxolan-3-yl]methyl}carbamoyl)phenyl]-1,3-dihydro-2H-isoindole-2-carboxamide, Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Longenecker, K.L, Raich, D, Korepanova, A.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery and Characterization of Novel Nonsubstrate and Substrate NAMPT Inhibitors.
Mol. Cancer Ther., 16, 2017
|
|
8AB5
 
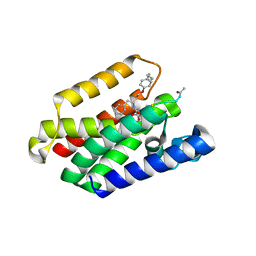 | | Structure of E. coli GlpG in complex with peptide derived inhibitor Ac-VRHA-conh-[4-(4-butyl)-phenoxy-1-phenyl-2-butyl] | | Descriptor: | Ac-VRHA-conh-[4-(4-butyl)-phenoxy-1-phenyl-2-butyl], Rhomboid protease GlpG | | Authors: | Skerlova, J, Polovinkin, V, Bach, K, Borshchevskiy, V, Strisovsky, K. | | Deposit date: | 2022-07-04 | | Release date: | 2023-10-25 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Extensive targeting of chemical space at the prime side of ketoamide inhibitors of rhomboid proteases by branched substituents empowers their selectivity and potency.
Eur.J.Med.Chem., 275, 2024
|
|
5K4C
 
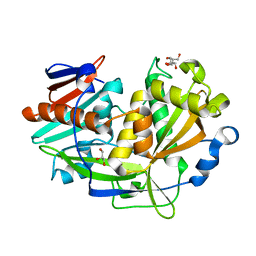 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 2 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit D, GLYCEROL | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
8ANT
 
 | |
7MLK
 
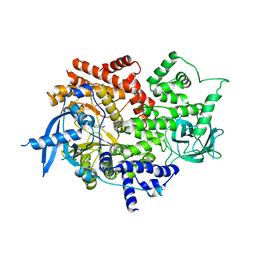 | | Crystal structure of human PI3Ka (p110a subunit) with MMV085400 bound to the active site determined at 2.9 angstroms resolution | | Descriptor: | 4-[6-(3,4,5-trimethoxyanilino)pyrazin-2-yl]benzamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Krake, S.H, Martinez, P.D.G, Poggi, M.L, Ferreira, M.S, Aguiar, A.C.C, Souza, G.E, Wenlock, M, Jones, B, Steinbrecher, T, Day, T, McPhail, J, Burke, J, Yeo, T, Mok, S, Uhlemann, A.C, Fidock, D.A, Chen, P, Grodsky, N, Deng, Y.L, Guido, R.V.C, Campbell, S.F, Willis, P.A, Dias, L.C. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Discovery of 2,6-disubstituted pyrazines as potent PI4K inhibitors with antimalarial activity
To Be Published
|
|
5K4M
 
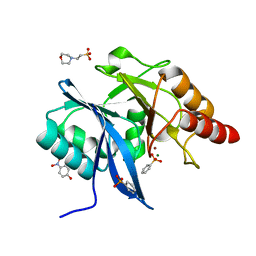 | |
5UD7
 
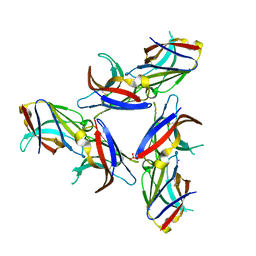 | | Crystal Structure of Wild-Type Ig-like Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, SULFATE ION, ... | | Authors: | Sudom, A, Min, X, Wang, Z. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.20002246 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
8IYV
 
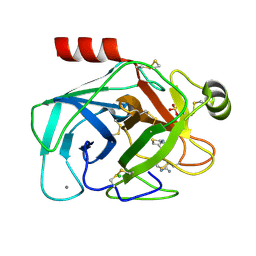 | | Crystal structure of trypsin-famotidine complex at 2.10 Angstroms resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cationic trypsin, ... | | Authors: | Ahmad, M.S, Kalam, N, Akbar, Z, Rasheed, S, Choudhary, M.I. | | Deposit date: | 2023-04-06 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the binding of famotidine, cimetidine, guanidine, and pimagedine with serine protease.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
5D5Y
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form I | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8IZK
 
 | | Crystal structure of trypsin-guanidine complex at 2.05 Angstroms resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cationic trypsin, ... | | Authors: | Ahmad, M.S, Kalam, N, Akbar, Z, Rasheed, S, Choudhary, M.I. | | Deposit date: | 2023-04-07 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the binding of famotidine, cimetidine, guanidine, and pimagedine with serine protease.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
5UDX
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with zinc | | Descriptor: | Lactate racemization operon protein LarE, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8IZH
 
 | | Crystal structure of trypsin-aminoguanidine complex at 2.30 Angstroms resolution | | Descriptor: | AMINOGUANIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ahmad, M.S, Kalam, N, Akbar, Z, Rasheed, S, Choudhary, M.I. | | Deposit date: | 2023-04-07 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the binding of famotidine, cimetidine, guanidine, and pimagedine with serine protease.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
7OML
 
 | |
