7JG2
 
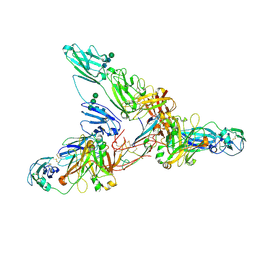 | | Secretory Immunoglobin A (SIgA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, ... | | Authors: | Kumar Bharathkar, S, Parker, B.P, Malyutin, A.G, Stadtmueller, B.M. | | Deposit date: | 2020-07-18 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structures of Secretory and dimeric Immunoglobulin A.
Elife, 9, 2020
|
|
3FVN
 
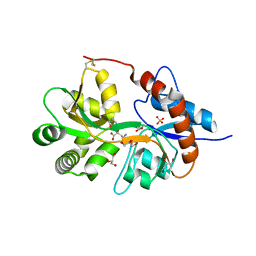 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with 9-deoxy-neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,7R,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-7-hydroxy-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
3FV2
 
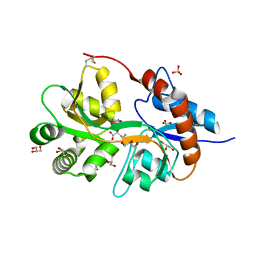 | | Crystal structure of the human glutamate receptor, GluR5, ligand-binding core in complex with neodysiherbaine A in space group P1 | | Descriptor: | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6,7-dihydroxyhexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
6DFR
 
 | |
3BKI
 
 | | Crystal Structure of the GluR2 ligand binding core (S1S2J) in complex with FQX at 1.87 Angstroms | | Descriptor: | Glutamate receptor 2, [1,2,5]oxadiazolo[3,4-g]quinoxaline-6,7(5H,8H)-dione 1-oxide | | Authors: | Cruz, L, Estebanez-Perpina, E, Pfaff, S, Borngraeber, S, Bao, N, Fletterick, R, England, P. | | Deposit date: | 2007-12-06 | | Release date: | 2008-09-16 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | 6-Azido-7-nitro-1,4-dihydroquinoxaline-2,3-dione (ANQX) forms an irreversible bond to the active site of the GluR2 AMPA receptor.
J.Med.Chem., 51, 2008
|
|
2QNZ
 
 | | Crystal structure of the complex between the mycobacterium beta-ketoacyl-acyl carrier protein synthase III (FABH) and SS-(2-hydroxyethyl)-O-decyl ester carbono(dithioperoxoic) acid | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, BETA-MERCAPTOETHANOL, DECYL FORMATE | | Authors: | Sachdeva, S, Musayev, F, Alhamadsheh, M, Scarsdale, J.N, Wright, H.T, Reynolds, K.A. | | Deposit date: | 2007-07-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Separate Entrance and Exit Portals for Ligand Traffic in Mycobacterium tuberculosis FabH
Chem.Biol., 15, 2008
|
|
8TZX
 
 | | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue dWIZ-1 | | Descriptor: | (3S)-3-(5-{(1R)-1-[(2R)-1-ethylpiperidin-2-yl]ethoxy}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, DNA damage-binding protein 1, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-28 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A molecular glue degrader of the WIZ transcription factor for fetal hemoglobin induction.
Science, 385, 2024
|
|
2QO0
 
 | | Crystal structure of the complex between the A246F mutant of mycobacterium beta-ketoacyl-acyl carrier protein synthase III (FABH) and 11-(decyldithiocarbonyloxy)-undecanoic acid | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, DECANE-1-THIOL | | Authors: | Sachdeva, S, Musayev, F, Alhamadsheh, M, Scarsdale, J.N, Wright, H.T, Reynolds, K.A. | | Deposit date: | 2007-07-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Separate Entrance and Exit Portals for Ligand Traffic in Mycobacterium tuberculosis FabH
Chem.Biol., 15, 2008
|
|
6L93
 
 | |
4WF2
 
 | | Structure of E. coli BirA G142A bound to biotinol-5'-AMP | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, Bifunctional ligase/repressor BirA | | Authors: | Eginton, C, Beckett, D, Wade, H. | | Deposit date: | 2014-09-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Allosteric Coupling via Distant Disorder-to-Order Transitions.
J.Mol.Biol., 427, 2015
|
|
6WN2
 
 | |
6WQ6
 
 | |
6WQS
 
 | | Xanthomonas citri Methionyl-tRNA synthetase in complex with REP8839 | | Descriptor: | 2-{[3-({[4-bromo-5-(1-fluoroethenyl)-3-methylthiophen-2-yl]methyl}amino)propyl]amino}quinolin-4(1H)-one, Methionine--tRNA ligase, ZINC ION | | Authors: | Mercaldi, G.F, Benedetti, C.E. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for diaryldiamine selectivity and competition with tRNA in a type 2 methionyl-tRNA synthetase from a Gram-negative bacterium.
J.Biol.Chem., 296, 2021
|
|
6WQT
 
 | | Xanthomonas citri Methionyl-tRNA synthetase in complex with REP3123 | | Descriptor: | 5-[(3-{[(4R)-6,8-dibromo-3,4-dihydro-2H-1-benzopyran-4-yl]amino}propyl)amino]thieno[3,2-b]pyridin-7(6H)-one, Methionine--tRNA ligase, ZINC ION | | Authors: | Mercaldi, G.F, Benedetti, C.E. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis for diaryldiamine selectivity and competition with tRNA in a type 2 methionyl-tRNA synthetase from a Gram-negative bacterium.
J.Biol.Chem., 296, 2021
|
|
3FV1
 
 | | Crystal Structure of the human glutamate receptor, GluR5, ligand-binding core in complex with dysiherbaine in space group P1 | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, GLYCEROL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2009-01-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
5LWJ
 
 | | Solution NMR structure of the GTP binding Class II RNA aptamer-ligand-complex containing a protonated adenine nucleotide with a highly shifted pKa. | | Descriptor: | GTP Class II RNA (34-MER), GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wolter, A.C, Weickhmann, A.K, Nasiri, A.H, Hantke, K, Ohlenschlaeger, O, Wunderlich, C.H, Kreutz, C, Duchardt-Ferner, E, Woehnert, J. | | Deposit date: | 2016-09-17 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Stably Protonated Adenine Nucleotide with a Highly Shifted pKa Value Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1PXD
 
 | | Crystal structure of the complex of jacalin with meso-tetrasulphonatophenylporphyrin. | | Descriptor: | 5,10,15,20-TETRAKIS(4-SULPFONATOPHENYL)-21H,23H-PORPHINE, Agglutinin alpha chain, Agglutinin beta-3 chain | | Authors: | Goel, M, Anuradha, P, Kaur, K.J, Maiya, B.G, Swamy, M.J, Salunke, D.M. | | Deposit date: | 2003-07-03 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Porphyrin binding to jacalin is facilitated by the inherent plasticity of the carbohydrate-binding site: novel mode of lectin-ligand interaction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6KXX
 
 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist (compound A) | | Descriptor: | 1-(4-chlorophenyl)-6-methyl-3-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, PGC1alpha, Peroxisome proliferator-activated receptor alpha | | Authors: | Yoshida, T, Tachibana, K, Oki, H, Doi, M, Fukuda, S, Yuzuriha, T, Tabata, R, Ishimoto, K, Kawahara, K, Ohkubo, T, Miyachi, H, Doi, T. | | Deposit date: | 2019-09-14 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for PPAR alpha Activation by 1H-pyrazolo-[3,4-b]pyridine Derivatives.
Sci Rep, 10, 2020
|
|
6KXY
 
 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist (compound B) | | Descriptor: | 6-ethyl-1-(4-fluorophenyl)-3-pentan-3-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, PGC1alpha, Peroxisome proliferator-activated receptor alpha | | Authors: | Yoshida, T, Tachibana, K, Oki, H, Doi, M, Fukuda, S, Yuzuriha, T, Tabata, R, Ishimoto, K, Kawahara, K, Ohkubo, T, Miyachi, H, Doi, T. | | Deposit date: | 2019-09-14 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for PPAR alpha Activation by 1H-pyrazolo-[3,4-b]pyridine Derivatives.
Sci Rep, 10, 2020
|
|
7BJV
 
 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI09181 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ecdysone Receptor, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Browning, C, McEwen, A.G, Billas, I.M.L. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Nonsteroidal ecdysone receptor agonists use a water channel for binding to the ecdysone receptor complex EcR/USP.
J Pestic Sci, 46, 2021
|
|
7BJU
 
 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI08346 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, Ecdysone Receptor, ... | | Authors: | Browning, C, McEwen, A.G, Billas, I.M.L. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Nonsteroidal ecdysone receptor agonists use a water channel for binding to the ecdysone receptor complex EcR/USP.
J Pestic Sci, 46, 2021
|
|
5HMK
 
 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {4-[2-(2-hydroxyethoxy)phenyl]piperazin-1-yl}[(2R,3S)-2-propyl-3-[4-(trifluoromethyl)phenoxy]-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}piperidin-3-yl]methanone | | Authors: | Scapin, G. | | Deposit date: | 2016-01-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
8J9Q
 
 | | Crystal structure of UBR box of UBR4 apo | | Descriptor: | E3 ubiquitin-protein ligase UBR4, ZINC ION | | Authors: | Jeong, D.-E, KIm, S.-J, Shin, H.-C. | | Deposit date: | 2023-05-04 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Insights into the recognition mechanism in the UBR box of UBR4 for its specific substrates.
Commun Biol, 6, 2023
|
|
8J9R
 
 | |
8V0E
 
 | | ANK repeat of MIB1 | | Descriptor: | E3 ubiquitin-protein ligase MIB1, Unidentified MIB1 peptide | | Authors: | Cao, R, Blacklow, S.C. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural requirements for activity of Mind bomb1 in Notch signaling.
Structure, 2024
|
|
