4ELF
 
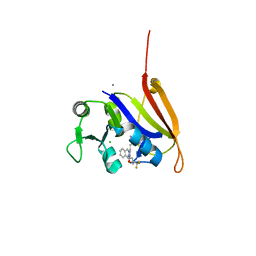 | | Structure-activity relationship guides enantiomeric preference among potent inhibitors of B. anthracis dihydrofolate reductase | | Descriptor: | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(3,3,3-trifluoropropyl)phthalazin-2(1H)-yl ]prop-2-en-1-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2012-04-10 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-activity relationship for enantiomers of potent inhibitors of B. anthracis dihydrofolate reductase.
Biochim.Biophys.Acta, 1834, 2013
|
|
2KE9
 
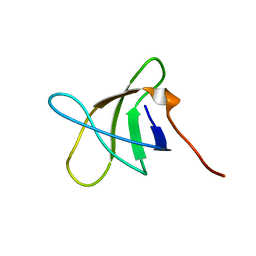 | |
2KZG
 
 | | A Transient and Low Populated Protein Folding Intermediate at Atomic Resolution | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Religa, T.L, Banachewicz, W, Fersht, A.R, Kay, L.E. | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A transient and low-populated protein-folding intermediate at atomic resolution.
Science, 329, 2010
|
|
2JBO
 
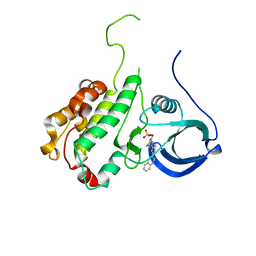 | | Protein kinase MK2 in complex with an inhibitor (crystal form-1, soaking) | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP KINASE-ACTIVATED PROTEIN KINASE 2, PHOSPHATE ION | | Authors: | Hillig, R.C, Eberspaecher, U, Monteclaro, F, Huber, M, Nguyen, D, Mengel, A, Muller-Tiemann, B, Egner, U. | | Deposit date: | 2006-12-09 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for a High Affinity Inhibitor Bound to Protein Kinase Mk2.
J.Mol.Biol., 369, 2007
|
|
2JBP
 
 | | Protein kinase MK2 in complex with an inhibitor (crystal form-2, co- crystallization) | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP KINASE-ACTIVATED PROTEIN KINASE 2 | | Authors: | Hillig, R.C, Eberspaecher, U, Monteclaro, F, Huber, M, Nguyen, D, Mengel, A, Muller-Tiemann, B, Egner, U. | | Deposit date: | 2006-12-09 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis for a high affinity inhibitor bound to protein kinase MK2.
J. Mol. Biol., 369, 2007
|
|
2JEV
 
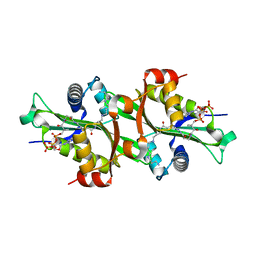 | | Crystal structure of human spermine,spermidine acetyltransferase in complex with a bisubstrate analog (N1-acetylspermine-S-CoA). | | Descriptor: | (3R)-27-AMINO-3-HYDROXY-2,2-DIMETHYL-4,8,14-TRIOXO-12-THIA-5,9,15,19,24-PENTAAZAHEPTACOS-1-YL [(2S,3R,4S,5S)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, DIAMINE ACETYLTRANSFERASE 1 | | Authors: | Hegde, S.S, Chandler, J, Vetting, M.W, Yu, M, Blanchard, J.S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic and Structural Analysis of Human Spermidine/Spermine N(1)-Acetyltransferase.
Biochemistry, 46, 2007
|
|
2JK8
 
 | |
2RFI
 
 | | Crystal structure of catalytic domain of human euchromatic histone methyltransferase 1 in complex with SAH and dimethylated H3K9 peptide | | Descriptor: | Histone H3, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Min, J, Wu, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-30 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
6SCZ
 
 | | Mycobacterium tuberculosis alanine racemase inhibited by DCS | | Descriptor: | (~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylidene-[(4~{R})-3-oxidanylidene-1,2-oxazolidin-4-yl]azanium, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | de Chiara, C, Purkiss, A, Prosser, G, Homsak, M, de Carvalho, L.P.S. | | Deposit date: | 2019-07-26 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | D-Cycloserine destruction by alanine racemase and the limit of irreversible inhibition.
Nat.Chem.Biol., 16, 2020
|
|
2JMO
 
 | | IBR domain of Human Parkin | | Descriptor: | Parkin, ZINC ION | | Authors: | Beasley, S.A, Hristova, V.A, Shaw, G.S. | | Deposit date: | 2006-11-24 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the Parkin in-between-ring domain provides insights for E3-ligase dysfunction in autosomal recessive Parkinson's disease.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2MKO
 
 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3'), POTASSIUM ION | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-11 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
2RAQ
 
 | | Crystal structure of the MTH889 protein from Methanothermobacter thermautotrophicus. Northeast Structural Genomics Consortium target TT205 | | Descriptor: | CALCIUM ION, Conserved protein MTH889 | | Authors: | Forouhar, F, Su, M, Xu, X, Seetharaman, J, Mao, L, Xiao, R, Ma, L.-C, Conover, K, Baran, M.C, Acton, T.B, Montelione, G.T, Arrowsmith, C.H, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-17 | | Release date: | 2007-10-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of the MTH889 protein from Methanothermobacter thermautotrophicus.
To be Published
|
|
6SAP
 
 | |
2OYH
 
 | | Crystal Structure of Fragment D of gammaD298,301A Fibrinogen with the Peptide Ligand Gly-His-Arg-Pro-Amide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha chain, ... | | Authors: | Kostelansky, M.S, Gorkun, O.V, Lord, S.T. | | Deposit date: | 2007-02-22 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the gamma2 Calcium-Binding Site: Studies with gammaD298,301A Fibrinogen Reveal Changes in the gamma294-301 Loop that Alter the Integrity of the "a" Polymerization Site.
Biochemistry, 46, 2007
|
|
2K36
 
 | | Structure ensemble Backbone and side-chain 1H, 13C, and 15N Chemical Shift Assignments, 1H-15N RDCs and 1H-1H nOe restraints for protein K7 from the Vaccinia virus | | Descriptor: | Protein K7 | | Authors: | Kalverda, A.P, Thompson, G.S, Vogel, A, Schr der, M, Bowie, A.G, Khan, A.R, Homans, S.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Poxvirus K7 protein adopts a Bcl-2 fold: biochemical mapping of its interactions with human DEAD box RNA helicase DDX3.
J.Mol.Biol., 385, 2009
|
|
2VF2
 
 | | X-ray crystal structure of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, SULFATE ION | | Authors: | Lack, N, Lowe, E.D, Liu, J, Eltis, L.D, Noble, M.E.M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Hsad, a Steroid-Degrading Hydrolase, from Mycobacterium Tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2KC9
 
 | |
2MKM
 
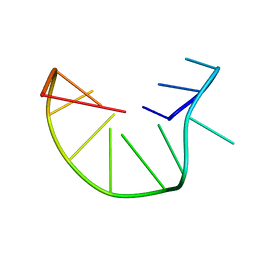 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3') | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-10 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
2MFZ
 
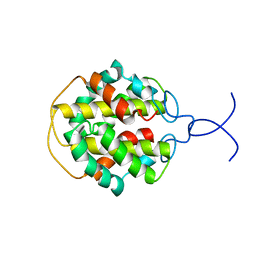 | | NMR structure of C-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Andersson, M, Chen, G, Landreh, M, Nordling, K, Kronqvist, N, Westermark, P, Jornvall, H, Knight, S, Ridderstrale, Y, Holm, L, Meng, Q, Chesler, M, Johansson, J, Rising, A. | | Deposit date: | 2013-10-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Carbonic Anhydrase Generates CO2 and H+ That Drive Spider Silk Formation Via Opposite Effects on the Terminal Domains
Plos Biol., 12, 2014
|
|
2LYG
 
 | | Fuc_TBA | | Descriptor: | 2-hydroxyethyl 6-deoxy-beta-L-galactopyranoside, DNA (5'-D(P*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avio, A, Eritja, R, Gonzalez-Ibaez, C, Morales, J. | | Deposit date: | 2012-09-18 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Carbohydrate-DNA interactions at G-quadruplexes: folding and stability changes by attaching sugars at the 5'-end.
Chemistry, 19, 2013
|
|
2L39
 
 | | Mouse prion protein fragment 121-231 AT 37 C | | Descriptor: | Major prion protein | | Authors: | Christen, B, Damberger, F.F, Perez, D.R, Hornemann, S, Wuthrich, K. | | Deposit date: | 2010-09-10 | | Release date: | 2011-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cellular prion protein conformation and function.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
9FEB
 
 | |
9B55
 
 | |
9GCJ
 
 | |
9EOQ
 
 | | Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA | | Descriptor: | DNA (42-MER), DNA (5'-D(P*AP*TP*AP*TP*AP*GP*CP*GP*TP*GP*GP*AP*AP*GP*T)-3') | | Authors: | Ali, K, Georg, K, Volodymyr, M, Johanna, G, Maximilian, N.H, Lukas, K, Simone, C, Hendrik, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Designing Rigid DNA Origami Templates for Molecular Visualization Using Cryo-EM.
Nano Lett., 24, 2024
|
|
