3GX7
 
 | |
3H5W
 
 | | Crystal structure of the GluR2-ATD in space group P212121 without solvent | | Descriptor: | Glutamate receptor 2 | | Authors: | Jin, R, Singh, S.K, Gu, S, Furukawa, H, Sobolevsky, A, Zhou, J, Jin, Y, Gouaux, E. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structure and association behaviour of the GluR2 amino-terminal domain.
Embo J., 28, 2009
|
|
3HAJ
 
 | | Crystal structure of human PACSIN2 F-BAR domain (p212121 lattice) | | Descriptor: | CALCIUM ION, human PACSIN2 F-BAR | | Authors: | Wang, Q, Navarro, M.V.A.S, Peng, G, Rajashankar, K.R, Sondermann, H. | | Deposit date: | 2009-05-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HAB
 
 | | The structure of DPP4 in complex with piperidine fused benzimidazole 25 | | Descriptor: | (2R,3R)-7-(methylsulfonyl)-3-(2,4,5-trifluorophenyl)-1,2,3,4-tetrahydropyrido[1,2-a]benzimidazol-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2009-05-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopiperidine-fused imidazoles as dipeptidyl peptidase-IV inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3HAW
 
 | |
3IJ9
 
 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | (2R,3S,4R,5R,6R)-2,6-difluoro-2-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
3IRW
 
 | | Structure of a c-di-GMP riboswitch from V. cholerae | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Smith, K.D. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding by a c-di-GMP riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
5NSK
 
 | | apo Structure of Leucyl aminopeptidase from Trypanosoma brucei | | Descriptor: | Aminopeptidase, putative | | Authors: | Timm, J, Wilson, K.S. | | Deposit date: | 2017-04-26 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2018
|
|
5NSD
 
 | | Co-crystal structure of NAMPT dimer with KPT-9274 | | Descriptor: | (~{E})-3-(6-azanylpyridin-3-yl)-~{N}-[[5-[4-[4,4-bis(fluoranyl)piperidin-1-yl]carbonylphenyl]-7-(4-fluorophenyl)-1-benzofuran-2-yl]methyl]prop-2-enamide, GLYCEROL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Neggers, J.E, Kwanten, B, Dierckx, T, Noguchi, H, Voet, A, Vercruysse, T, Baloglu, E, Senapedis, W, Jacquemyn, M, Daelemans, D. | | Deposit date: | 2017-04-26 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Target identification of small molecules using large-scale CRISPR-Cas mutagenesis scanning of essential genes.
Nat Commun, 9, 2018
|
|
5O2R
 
 | | Cryo-EM structure of the proline-rich antimicrobial peptide Api137 bound to the terminating ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5NXN
 
 | |
5NP6
 
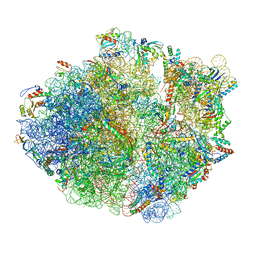 | | 70S structure prior to bypassing | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Klimova, M, Zamora, M, Gil-Carton, D, Rodnina, M, Valle, M. | | Deposit date: | 2017-04-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ribosome rearrangements at the onset of translational bypassing.
Sci Adv, 3, 2017
|
|
3I6W
 
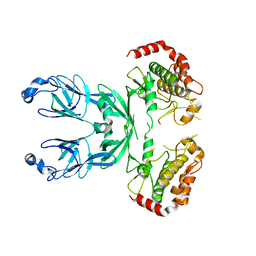 | |
3IAR
 
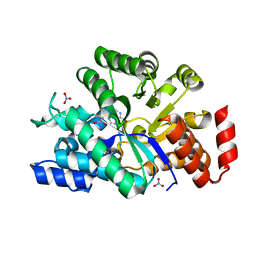 | | The crystal structure of human adenosine deaminase | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, Adenosine deaminase, GLYCEROL, ... | | Authors: | Ugochukwu, E, Zhang, Y, Hapka, E, Yue, W.W, Bray, J.E, Muniz, J, Burgess-Brown, N, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-14 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The crystal structure of human adenosine deaminase
To be Published
|
|
5N10
 
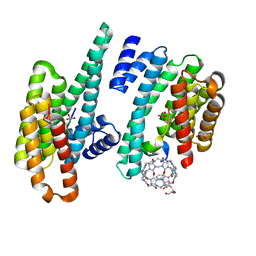 | |
5NHL
 
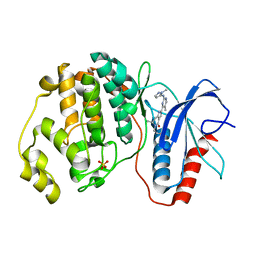 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | (6~{R})-5-(2-methoxyethyl)-6-methyl-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NMO
 
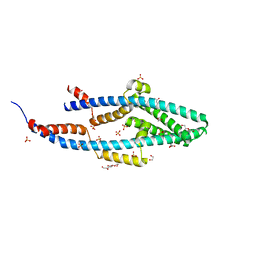 | | Structure of the Bacillus subtilis Smc Joint domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Chromosome partition protein Smc,Chromosome partition protein Smc, ... | | Authors: | Diebold-Durand, M.-L, Basquin, J, Gruber, S. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization.
Mol. Cell, 67, 2017
|
|
5NO4
 
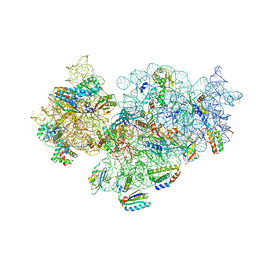 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate with uS3) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
5NO2
 
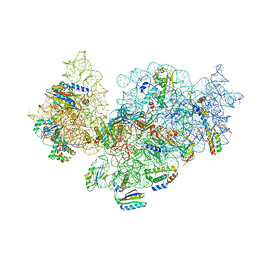 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
5NPY
 
 | | Crystal structure of Helicobacter pylori flagellar hook protein FlgE2 | | Descriptor: | Flagellar basal body protein, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Loconte, V, Zanotti, G, Kekez, I, Matkovic-Calogovic, D. | | Deposit date: | 2017-04-19 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural characterization of FlgE2 protein from Helicobacter pylori hook.
FEBS J., 284, 2017
|
|
5N7E
 
 | | Crystal structure of the Dbl-homology domain of Bcr-Abl in complex with monobody Mb(Bcr-DH_4). | | Descriptor: | Breakpoint cluster region protein, Mb(Bcr-DH_4) | | Authors: | Reckel, S, Reynaud, A, Pojer, F, Hantschel, O. | | Deposit date: | 2017-02-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
3GX5
 
 | |
5Q0C
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(4-methoxyphenyl)thiophen-2-yl]sulfonylurea and with fructose-2,6-diphosphate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(4-methoxyphenyl)thiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(4-methoxyphenyl)thiophen-2-yl]sulfonylurea and with f2,6p
To be published
|
|
3H2W
 
 | |
3I6Z
 
 | | 3D Structure of Torpedo californica acetylcholinesterase complexed with N-saccharinohexyl-galanthamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{6-[(4aS,6R,8aS)-6-hydroxy-3-methoxy-5,6,9,10-tetrahydro-4aH-[1]benzofuro[3a,3,2-ef][2]benzazepin-11(12H)-yl]hexyl}-1 ,2-benzisothiazol-3(2H)-one 1,1-dioxide, ... | | Authors: | Lamba, D, Bartolucci, C. | | Deposit date: | 2009-07-07 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Probing Torpedo californica acetylcholinesterase catalytic gorge with two novel bis-functional galanthamine derivatives.
J.Med.Chem., 53, 2010
|
|
