9B8W
 
 | |
8WRL
 
 | | XBB.1.5 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
9F6F
 
 | | Human DNA polymerase epsilon bound to DNA and PCNA (closed conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 2024
|
|
9B8Y
 
 | |
9B8X
 
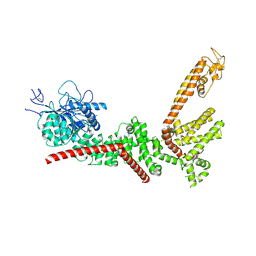 | |
9F6K
 
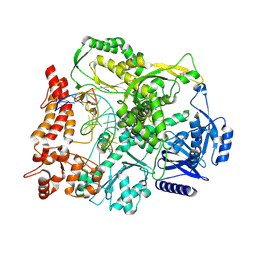 | |
8WRM
 
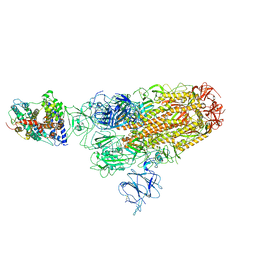 | | XBB.1.5 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRO
 
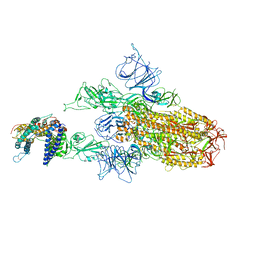 | | XBB.1.5.10 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein,Spike glycoprotein,Spike glycoprotein,Fusion protein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTD
 
 | | XBB.1.5.10 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTJ
 
 | | XBB.1.5.70 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
9F6E
 
 | | Human DNA polymerase epsilon bound to DNA and PCNA (ajar conformation) | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA nascent strand, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Roske, J.J, Yeeles, J.T.P. | | Deposit date: | 2024-05-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Nat.Struct.Mol.Biol., 2024
|
|
8YT4
 
 | | Structure of Aquifex aeolicus Lumazine Synthase by Cryo-Electron Microscopy to 1.42 Angstrom Resolution | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Savva, C.G, Sobhy, M.A, De Biasio, A, Hamdan, S.M. | | Deposit date: | 2024-03-24 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (1.42 Å) | | Cite: | Structure of Aquifex aeolicus lumazine synthase by cryo-electron microscopy to 1.42 angstrom resolution.
Iucrj, 2024
|
|
8P6Y
 
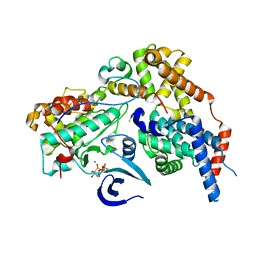 | | Cryo-EM structure of CAK in complex with nucleotide analogue ATPgS | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P7W
 
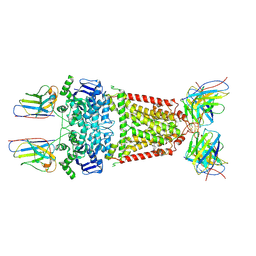 | | Structure of 5D3-Fab and nanobody(Nb8)-bound ABCG2 | | Descriptor: | 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ATP-binding cassette sub-family G member 2, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-05-31 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
8P8J
 
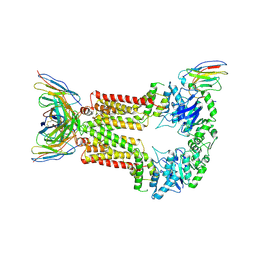 | | Structure of 5D3-Fab and nanobody(Nb96)-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-06-01 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
9B94
 
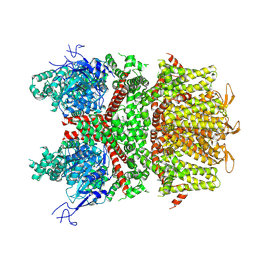 | |
7RW6
 
 | | BORF2-APOBEC3Bctd Complex | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3B, Maltose/maltodextrin-binding periplasmic protein,Ribonucleoside-diphosphate reductase large subunit, ZINC ION | | Authors: | Shaban, N.M, Yan, R, Shi, K, McLellan, J.S, Yu, Z, Harris, R.S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure of the EBV ribonucleotide reductase BORF2 and mechanism of APOBEC3B inhibition.
Sci Adv, 8, 2022
|
|
8P6V
 
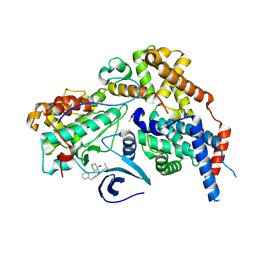 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0942 | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
9B91
 
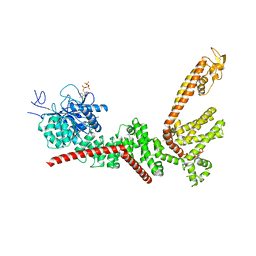 | |
8ORM
 
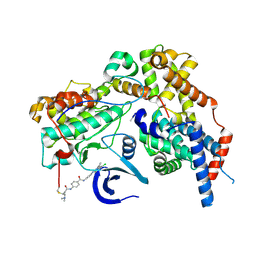 | | Cryo-EM structure of CAK-THZ1 | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P8A
 
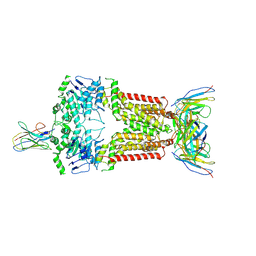 | | Structure of 5D3-Fab and nanobody(Nb17)-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-05-31 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
7RU1
 
 | | SARS-CoV-2-6P-Mut7 S protein (C3 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU4
 
 | | CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.33 IgG heavy chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU8
 
 | | CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.30 Fab Kappa chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
5E4M
 
 | |
