5B6G
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
2BB5
 
 | | Structure of Human Transcobalamin in complex with Cobalamin | | Descriptor: | COBALAMIN, Transcobalamin II | | Authors: | Wuerges, J, Garau, G, Geremia, S, Fedosov, S.N, Petersen, T.E, Randaccio, L. | | Deposit date: | 2005-10-17 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for mammalian vitamin B12 transport by transcobalamin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2BBC
 
 | | Structure of Cobalamin-complexed Bovine Transcobalamin in trigonal crystal form | | Descriptor: | CHLORIDE ION, COBALAMIN, Transcobalamin II | | Authors: | Wuerges, J, Garau, G, Geremia, S, Fedosov, S.N, Petersen, T.E, Randaccio, L. | | Deposit date: | 2005-10-17 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for mammalian vitamin B12 transport by transcobalamin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3QDL
 
 | | Crystal structure of RdxA from Helicobacter pyroli | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Oxygen-insensitive NADPH nitroreductase | | Authors: | Rojas, A.L, Martinez-Julvez, M, Olekhnovich, I.N, Hoffman, P.S, Sancho, J. | | Deposit date: | 2011-01-18 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of RdxA--an oxygen-insensitive nitroreductase essential for metronidazole activation in Helicobacter pylori.
Febs J., 279, 2012
|
|
4PXK
 
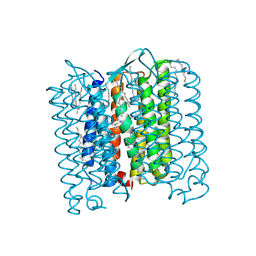 | | Crystal structure of Haloarcula marismortui bacteriorhodopsin I D94N mutant | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Shevchenko, V, Gushchin, I, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2014-03-24 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Escherichia coli-Expressed Haloarcula marismortui Bacteriorhodopsin I in the Trimeric Form.
Plos One, 9, 2014
|
|
1HPB
 
 | |
3HU2
 
 | | Structure of p97 N-D1 R86A mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.-K. | | Deposit date: | 2009-06-12 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
4QC9
 
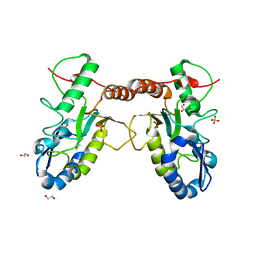 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uracil-DNA glycosylase | | Authors: | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.259 Å) | | Cite: | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4
To be Published
|
|
4EWA
 
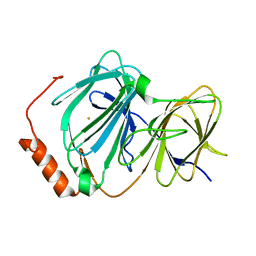 | | Study on structure and function relationships in human Pirin with Fe ion | | Descriptor: | FE (III) ION, Pirin | | Authors: | Liu, F, Rehmani, I, Chen, L, Fu, R, Serrano, V, Wilson, D.W, Liu, A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Pirin is an iron-dependent redox regulator of NF-kappa B.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5D1O
 
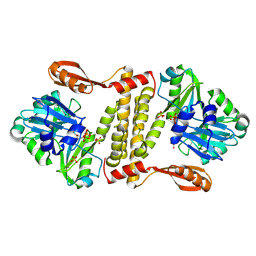 | | Archaeal ATP-dependent RNA ligase - form 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA ligase, MAGNESIUM ION, ... | | Authors: | Murakami, K.S. | | Deposit date: | 2015-08-04 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Structural and mutational analysis of archaeal ATP-dependent RNA ligase identifies amino acids required for RNA binding and catalysis.
Nucleic Acids Res., 44, 2016
|
|
4ERV
 
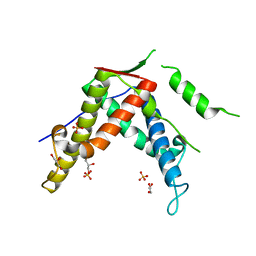 | |
3RFU
 
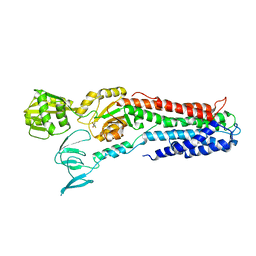 | | Crystal structure of a copper-transporting PIB-type ATPase | | Descriptor: | Copper efflux ATPase, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Gourdon, P, Liu, X, Skjorringe, T, Morth, J.P, Birk Moller, L, Panyella Pedersen, B, Nissen, P. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a copper-transporting PIB-type ATPase.
Nature, 475, 2011
|
|
2ZNX
 
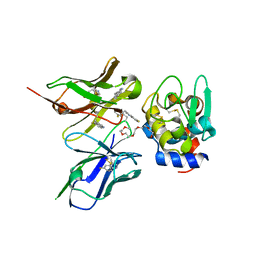 | | 5-Fluorotryptophan Incorporated ScFv10 Complexed to Hen Egg Lysozyme | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Lysozyme C, ScFv | | Authors: | DeSantis, M.E, Acchione, M, Li, M, Walter, R.L, Wlodawer, A, Smith-Gill, S. | | Deposit date: | 2008-05-02 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specific fluorine labeling of the HyHEL10 antibody affects antigen binding and dynamics
Biochemistry, 51, 2012
|
|
3TL8
 
 | |
3LFP
 
 | |
5D71
 
 | | Crystal structure of MOR04302, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | DI(HYDROXYETHYL)ETHER, Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
4ZBO
 
 | | Streptomyces bingchenggensis acetoacetate decarboxylase in non-covalent complex with potassium formate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Acetoacetate decarboxylase, ... | | Authors: | Mydy, L.S, Silvaggi, N.R. | | Deposit date: | 2015-04-15 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sbi00515, a Protein of Unknown Function from Streptomyces bingchenggensis, Highlights the Functional Versatility of the Acetoacetate Decarboxylase Scaffold.
Biochemistry, 54, 2015
|
|
4Q2G
 
 | | CRYSTAL STRUCTURE OF AN INTRAMEMBRANE CDP-DAG SYNTHETASE CENTRAL FOR PHOSPHOLIPID BIOSYNTHESIS (S200C/S223C, inactive mutant) | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, Phosphatidate cytidylyltransferase, ... | | Authors: | Liu, X, Yin, Y, Wu, J, Liu, Z. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis
Nat Commun, 5, 2014
|
|
1HTL
 
 | |
4ZGL
 
 | | Hit Like Protein | | Descriptor: | ADENOSINE MONOPHOSPHATE, Uncharacterized HIT-like protein HP_0404 | | Authors: | Tarique, K.F, Devi, S, Abdul Rehman, S.A, Gourinath, S. | | Deposit date: | 2015-04-23 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of HINT from Helicobacter pylori.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3CD3
 
 | | Crystal structure of phosphorylated human feline sarcoma viral oncogene homologue (v-FES) in complex with staurosporine and a consensus peptide | | Descriptor: | CHLORIDE ION, Proto-oncogene tyrosine-protein kinase Fes/Fps, STAUROSPORINE, ... | | Authors: | Filippakopoulos, P, Salah, E, Cooper, C, Picaud, S.S, Elkins, J.M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Coupling of SH2-Kinase Domains Links Fes and Abl Substrate Recognition and Kinase Activation
Cell(Cambridge,Mass.), 134, 2008
|
|
4TOI
 
 | | Crystal structure of E.coli ribosomal protein S2 in complex with N-terminal domain of S1 | | Descriptor: | 30S ribosomal protein S2,Ribosomal protein S1, ZINC ION | | Authors: | Grishkovskaya, I, Byrgazov, K, Moll, I, Djinovic-Carugo, K. | | Deposit date: | 2014-06-05 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the interaction of protein S1 with the Escherichia coli ribosome.
Nucleic Acids Res., 43, 2015
|
|
3CAL
 
 | |
3CBL
 
 | | Crystal structure of human feline sarcoma viral oncogene homologue (v-FES) in complex with staurosporine and a consensus peptide | | Descriptor: | Proto-oncogene tyrosine-protein kinase Fes/Fps, STAUROSPORINE, Synthetic peptide | | Authors: | Filippakopoulos, P, Salah, E, Cooper, C, Picaud, S.S, Elkins, J.M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-22 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Coupling of SH2-Kinase Domains Links Fes and Abl Substrate Recognition and Kinase Activation
Cell(Cambridge,Mass.), 134, 2008
|
|
4DJ2
 
 | | Unwinding the Differences of the Mammalian PERIOD Clock Proteins from Crystal Structure to Cellular Function | | Descriptor: | Period circadian protein homolog 1 | | Authors: | Kucera, N, Schmalen, I, Hennig, S, Oellinger, R, Strauss, H.M, Grudziecki, A, Wieczorek, C, Kramer, A, Wolf, E. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Unwinding the differences of the mammalian PERIOD clock proteins from crystal structure to cellular function.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
