2G7P
 
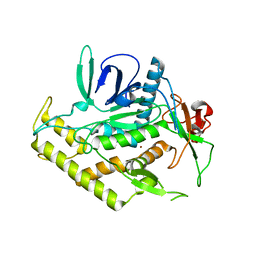 | | Structure of the Light Chain of Botulinum Neurotoxin Serotype A Bound to Small Molecule Inhibitors | | Descriptor: | Botulinum neurotoxin type A, ZINC ION | | Authors: | Fu, Z, Baldwin, M.R, Boldt, G.E, Janda, K.D, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2006-02-28 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Light chain of botulinum neurotoxin serotype A: structural resolution of a catalytic intermediate.
Biochemistry, 45, 2006
|
|
2G7K
 
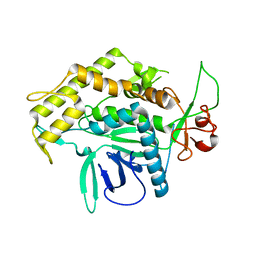 | | Structure of the Light Chain of Botulinum Neurotoxin, Serotype A Bound to small Molecule Inhibitors | | Descriptor: | Botulinum neurotoxin type A | | Authors: | Fu, Z, Baldwin, M.R, Boldt, G.E, Crawford, A, Janda, K.D, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2006-02-28 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Light chain of botulinum neurotoxin serotype A: structural resolution of a catalytic intermediate.
Biochemistry, 45, 2006
|
|
2G7Q
 
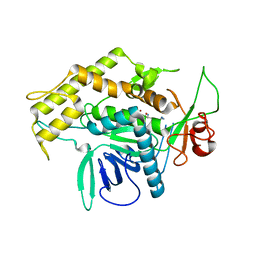 | | Structure of the Light Chain of Botulinum Neurotoxin Serotype A Bound to Small Molecule Inhibitors | | Descriptor: | Botulinum neurotoxin type A, N-HYDROXY-L-ARGININAMIDE, ZINC ION | | Authors: | Fu, Z, Baldwin, M.R, Boldt, G.E, Janda, K.D, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2006-02-28 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Light chain of botulinum neurotoxin serotype A: structural resolution of a catalytic intermediate.
Biochemistry, 45, 2006
|
|
4RF1
 
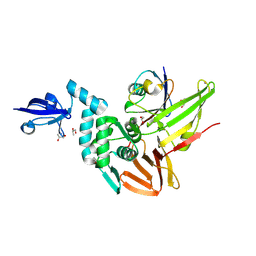 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P63) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, S-1,2-PROPANEDIOL, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
4REZ
 
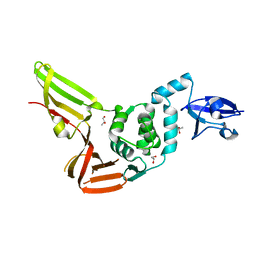 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease | | Descriptor: | ORF1ab protein, S-1,2-PROPANEDIOL, ZINC ION | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
4RF0
 
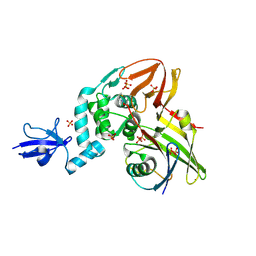 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P6522) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, SULFATE ION, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
1ELV
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COMPLEMENT C1S PROTEASE | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C1S COMPONENT, ... | | Authors: | Gaboriaud, C, Rossi, V, Bally, I, Arlaud, G, Fontecilla-Camps, J.-C. | | Deposit date: | 2000-03-14 | | Release date: | 2001-03-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the catalytic domain of human complement c1s: a serine protease with a handle.
EMBO J., 19, 2000
|
|
1EHF
 
 | |
1OGO
 
 | | Dex49A from Penicillium minioluteum complex with isomaltose | | Descriptor: | DEXTRANASE, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Larsson, A.M, Stahlberg, J, Jones, T.A. | | Deposit date: | 2003-05-08 | | Release date: | 2003-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dextranase from Penicillium Minioluteum. Reaction Course, Crystal Structure, and Product Complex
Structure, 11, 2003
|
|
7BSU
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in PtdSer-bound E2BeF state | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7BSW
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in PtdEtn-occluded E2-AlF state | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7CQY
 
 | |
6QJJ
 
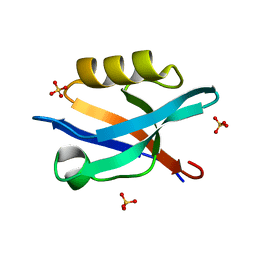 | |
6TA7
 
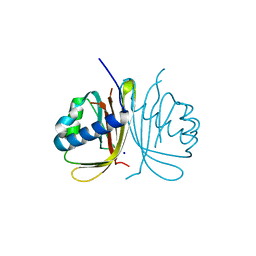 | | CRYSTAL STRUCTURE OF HUMAN G3BP1-NTF2 IN COMPLEX WITH HUMAN CAPRIN1-DERIVED SOLOMON MOTIF | | Descriptor: | CHLORIDE ION, Caprin-1, Ras GTPase-activating protein-binding protein 1, ... | | Authors: | Schulte, T, Achour, A, Panas, M.D, McInerney, G.M. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Caprin-1 binding to the critical stress granule protein G3BP1 is regulated by pH
Biorxiv, 2021
|
|
6QJD
 
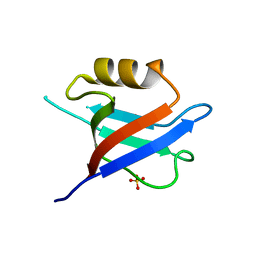 | |
6QJN
 
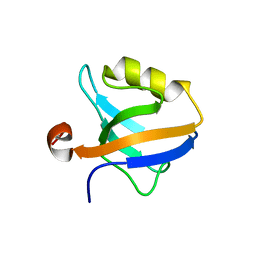 | |
6QJI
 
 | |
6QJF
 
 | |
6QJL
 
 | |
6QJG
 
 | |
6QJK
 
 | |
1OGM
 
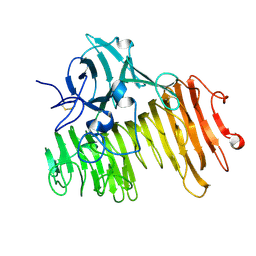 | |
5XA0
 
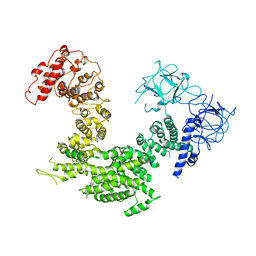 | | Crystal structure of inositol 1,4,5-trisphosphate receptor cytosolic domain | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Hamada, K, Miyatake, H, Terauchi, A, Mikoshiba, K. | | Deposit date: | 2017-03-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (5.812 Å) | | Cite: | IP3-mediated gating mechanism of the IP3 receptor revealed by mutagenesis and X-ray crystallography
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6DQV
 
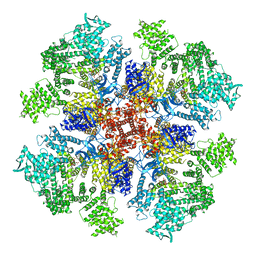 | | Class 2 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5XA1
 
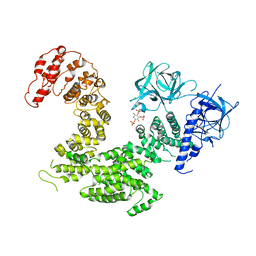 | | Crystal structure of inositol 1,4,5-trisphosphate receptor cytosolic domain with inositol 1,4,5-trisphosphate | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Hamada, K, Miyatake, H, Terauchi, A, Mikoshiba, K. | | Deposit date: | 2017-03-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (6.204 Å) | | Cite: | IP3-mediated gating mechanism of the IP3 receptor revealed by mutagenesis and X-ray crystallography
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
