5BQ8
 
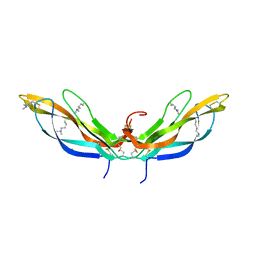 | | Crystal structure of Norrin, a Wnt signalling activator, Crystal Form II | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Norrin | | Authors: | Chang, T.-H, Hsieh, F.-L, Harlos, K, Jones, E.Y. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan.
Elife, 4, 2015
|
|
8J7E
 
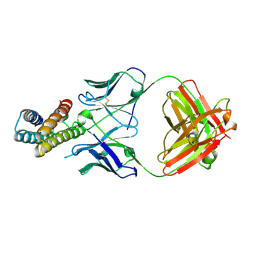 | | Crystal structure of BRIL in complex with 1b3 Fab | | Descriptor: | Antibody 1b3 Fab Heavy chain, Antibody 1b3 Fab Light chain, Soluble cytochrome b562 | | Authors: | Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8J3P
 
 | | Formate dehydrogenase mutant from from Candida dubliniensis M4 complexed with NADP+ | | Descriptor: | Formate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ma, W, Zheng, Y.C, Geng, Q, Chen, C, Xu, J.H. | | Deposit date: | 2023-04-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering a Formate Dehydrogenase for NADPH Regeneration.
Chembiochem, 24, 2023
|
|
8J69
 
 | |
5BS0
 
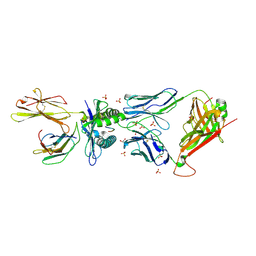 | | MAGE-A3 Reactive TCR in complex with Titin Epitope in HLA-A1 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Raman, M.C.C, Rizkallah, P.J, Simmons, R, Donellan, Z, Dukes, J, Bossi, G, LeProvost, G, Mahon, T, Hickman, E, Lomax, M, Oates, J, Hassan, N, Vuidepot, A, Sami, M, Cole, D.K, Jakobsen, B.K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Direct molecular mimicry enables off-target cardiovascular toxicity by an enhanced affinity TCR designed for cancer immunotherapy.
Sci Rep, 6, 2016
|
|
8J1X
 
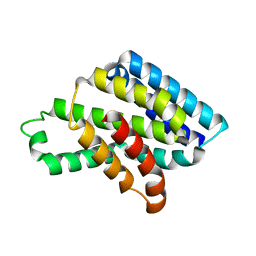 | |
8J1W
 
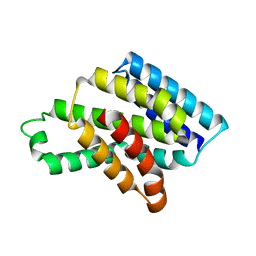 | |
5BV7
 
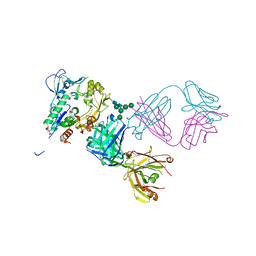 | | Crystal structure of human LCAT (L4F, N5D) in complex with Fab of an agonistic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 27C3 heavy chain, 27C3 light chain, ... | | Authors: | Piper, D.E, Romanow, W.G, Thibault, S.T, Walker, N.P.C. | | Deposit date: | 2015-06-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Agonistic Human Antibodies Binding to Lecithin-Cholesterol Acyltransferase Modulate High Density Lipoprotein Metabolism.
J.Biol.Chem., 291, 2016
|
|
5APJ
 
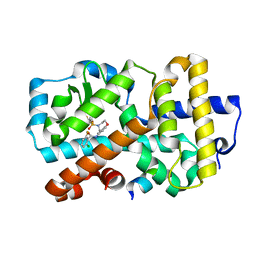 | | Ligand complex of RORg LBD | | Descriptor: | 2-CHLORO-6-FLUORO-N-[4-[3-(TRIFLUOROMETHYL)PHENYL]SULFONYL-3,5-DIHYDRO-2H-1,4-BENZOXAZEPIN-7-YL]BENZAMIDE, NUCLEAR RECEPTOR COACTIVATOR 2, NUCLEAR RECEPTOR ROR-GAMMA, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2015-09-16 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Benzoxazepines Achieve Potent Suppression of IL-17 Release in Human T-Helper 17 (TH 17) Cells through an Induced-Fit Binding Mode to the Nuclear Receptor ROR gamma.
ChemMedChem, 11, 2016
|
|
5BNC
 
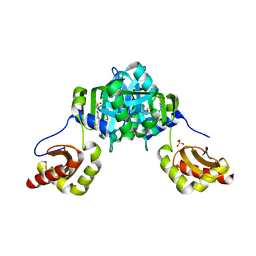 | | Structure of heme binding protein MSMEG_6519 from Mycobacterium smegmatis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Ahmed, F.H, Carr, P.D, Jackson, C.J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sequence-Structure-Function Classification of a Catalytically Diverse Oxidoreductase Superfamily in Mycobacteria.
J.Mol.Biol., 427, 2015
|
|
5BO7
 
 | | Structure of human sialyltransferase ST8SiaIII in complex with CTP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Volkers, G, Worrall, L, Strynadka, N.C.J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5ARM
 
 | | APO-CSP3 (COPPER STORAGE PROTEIN 3) FROM METHYLOSINUS | | Descriptor: | CSP3 | | Authors: | Vita, N, Landolfi, G, Basle, A, Platsaki, S, Waldron, K, Dennison, C. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.194 Å) | | Cite: | Novel Cytosolic Copper Storage Proteins
To be Published
|
|
5AMV
 
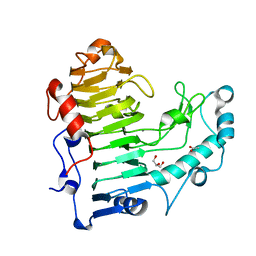 | | Structural insights into the loss of catalytic competence in pectate lyase at low pH | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Teixeira, S.C.M, Ali, S, Sondergaard, C, Pickersgill, R. | | Deposit date: | 2015-09-02 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Insights Into the Loss of Catalytic Competence in Pectate Lyase Activity at Low Ph.
FEBS Lett., 589, 2015
|
|
8J25
 
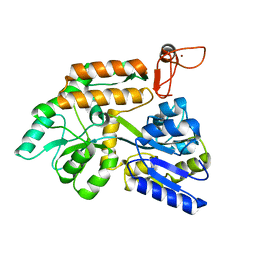 | | Crystal structure of PML B-box2 mutant | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
5BPB
 
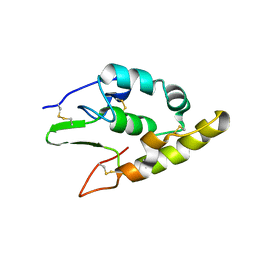 | | Crystal structure of the cysteine-rich domain of human Frizzled 4 - Crystal Form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4 | | Authors: | Chang, T.-H, Hsieh, F.-L, Harlos, K, Jones, E.Y. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan.
Elife, 4, 2015
|
|
5BQN
 
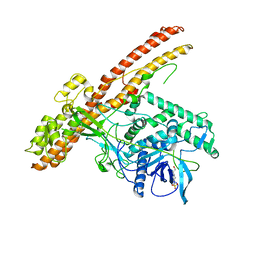 | | Crystal structure of the LHn fragment of botulinum neurotoxin type D, mutant H233Y E230Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Botulinum neurotoxin type D,Botulinum neurotoxin type D | | Authors: | Masuyer, G, Davies, J.R, Moore, K, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of Clostridium botulinum neurotoxin type D as a platform for the development of targeted secretion inhibitors.
Sci Rep, 5, 2015
|
|
5BRV
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, ZINC ION, pentamethylcyclopentadienyl iridium [N-benzensulfonamide-(2-pyridylmethyl-4-benzensulfonamide)amin] chloride | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
8JDN
 
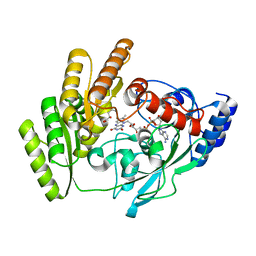 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxyvaleric acid | | Descriptor: | (2R)-2-oxidanylpentanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDS
 
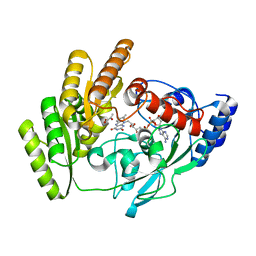 | | Crystal structure of mLDHD in complex with Pyruvate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.636 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8J98
 
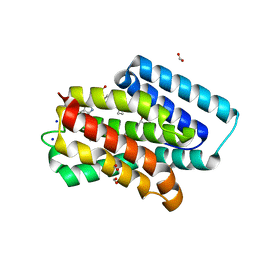 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | De novo backbone design for monomerization of near-infrared fluorescent protein
To Be Published
|
|
5BTT
 
 | | Switching GFP fluorescence using genetically encoded phenyl azide chemistry through two different non-native post-translational modifications routes at the same position. | | Descriptor: | GLYCEROL, Green fluorescent protein, SULFATE ION | | Authors: | Hartley, A.M, Worthy, H.L, Reddington, S.C, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
8JDD
 
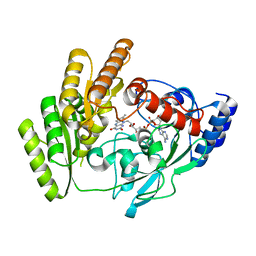 | | Crystal structure of H405A mLDHD in apo form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, mitochondrial | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDQ
 
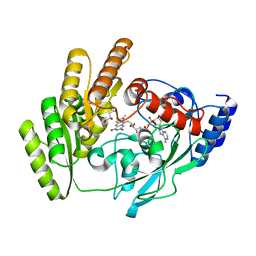 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxyisocaproic acid | | Descriptor: | (2R)-2-hydroxy-4-methylpentanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDZ
 
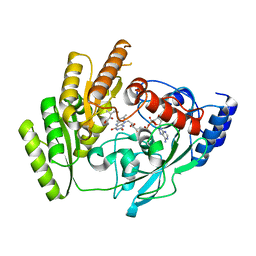 | | Crystal structure of mLDHD in complex with 2-keto-3-methylvaleric acid | | Descriptor: | (3S)-3-methyl-2-oxopentanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JEA
 
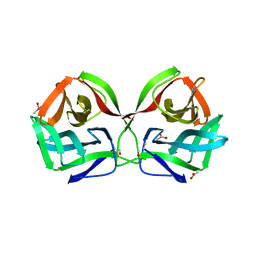 | | Crystal structure of CGL1 from Crassostrea gigas, mannotriose-bound form (CGL1/Man(alpha)1-2Man(alpha)1-2Man) | | Descriptor: | ACETIC ACID, CACODYLATE ION, MAGNESIUM ION, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Mannose oligosaccharide recognition of CGL1, a mannose-specific lectin containing DM9 motifs from Crassostrea gigas, revealed by X-ray crystallographic analysis.
J.Biochem., 175, 2023
|
|
