9B70
 
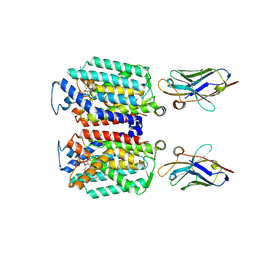 | | Cryo-EM structure of MraY in complex with analogue 2 | | Descriptor: | (2~{S},3~{S})-3-[(2~{S},3~{R},4~{S},5~{R})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-2-[[3-[[[(2~{S})-6-azanyl-2-(hexadecanoylamino)hexanoyl]amino]methyl]phenyl]methylamino]-3-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propanoic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2024-03-26 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Development of a natural product optimization strategy for inhibitors against MraY, a promising antibacterial target.
Nat Commun, 15, 2024
|
|
9B22
 
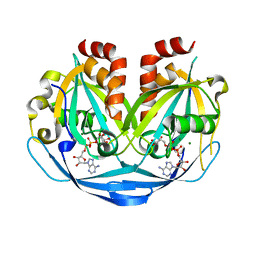 | |
9B0M
 
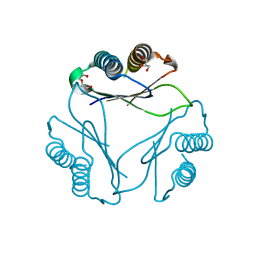 | |
9AZZ
 
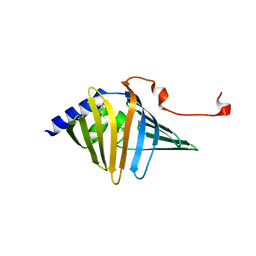 | |
9AYF
 
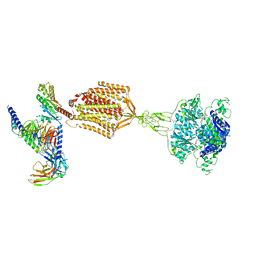 | | Structure of human calcium-sensing receptor in complex with Gi1 (miniGi1) protein in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AYA
 
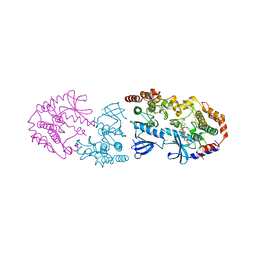 | | Crystal structure of CRAF/MEK complex with NST-628 and active RAF dimer | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, N-[3-fluoro-4-({7-[(3-fluoropyridin-2-yl)oxy]-4-methyl-2-oxo-2H-1-benzopyran-3-yl}methyl)pyridin-2-yl]-N'-methylsulfuric diamide, ... | | Authors: | Quade, B, Huang, X. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AY7
 
 | | Crystal structure of CRAF/MEK1 complex with NST-628 and inactive RAF | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Quade, B, Huang, X. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXY
 
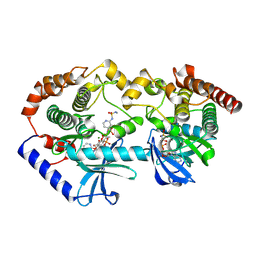 | | Crystal structure of BRAF/MEK complex with NST-628 and inactive RAF | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, N-[3-fluoro-4-({7-[(3-fluoropyridin-2-yl)oxy]-4-methyl-2-oxo-2H-1-benzopyran-3-yl}methyl)pyridin-2-yl]-N'-methylsulfuric diamide, ... | | Authors: | Quade, B, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXX
 
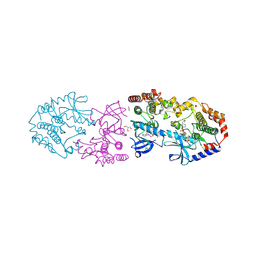 | | Crystal structure of BRAF/MEK1 complex with NST-628 and an active RAF dimer | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Quade, B, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXM
 
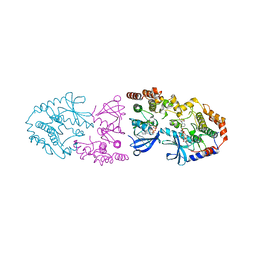 | | Crystal structure of ARAF/MEK1 complex with NST-628 and a RAF dimer | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 1, GLYCEROL, ... | | Authors: | Quade, B, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXH
 
 | | Crystal structure of KSR1/MEK1 complex heterotetramer with NST-628 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Quade, B, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXF
 
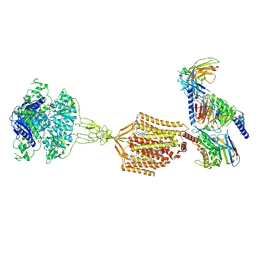 | | Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AXC
 
 | | Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, GST26/CRAF chimera, N-[3-fluoro-4-({7-[(3-fluoropyridin-2-yl)oxy]-4-methyl-2-oxo-2H-1-benzopyran-3-yl}methyl)pyridin-2-yl]-N'-methylsulfuric diamide | | Authors: | Quade, B, Cohen, S.E, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXA
 
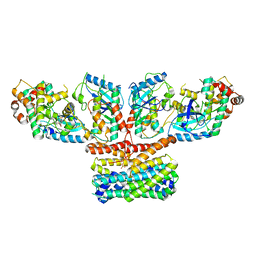 | | CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628 | | Descriptor: | 14-3-3 protein sigma, Dual specificity mitogen-activated protein kinase kinase 1, GST26/CRAF chimera, ... | | Authors: | Quade, B, Cohen, S.E, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AX6
 
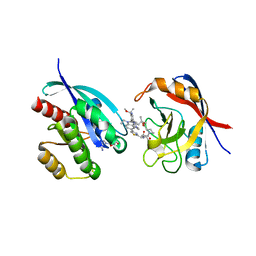 | | Tricomplex of RMC-6236, KRAS G12D, and CypA | | Descriptor: | (1R,2S)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1R)-1-methoxyethyl]-5-(4-methylpiperazin-1-yl)pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-2-methylcyclopropane-1-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Translational and Therapeutic Evaluation of RAS-GTP Inhibition by RMC-6236 in RAS-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AVQ
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9AVL
 
 | | Structure of human calcium-sensing receptor in complex with Gi3 protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AVK
 
 | | Structure of long Rib domain from Limosilactobacillus reuteri | | Descriptor: | SODIUM ION, THIOCYANATE ION, YSIRK signal domain/LPXTG anchor domain surface protein | | Authors: | Xue, Y, Kang, X. | | Deposit date: | 2024-03-04 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structure of the long Rib domain of the LPXTG-anchored surface protein from Limosilactobacillus reuteri.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9AVG
 
 | | Structure of human calcium-sensing receptor in complex with chimeric Gs (miniGis) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-02 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AVA
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | (2R)-2-[(5R,6S,8R,9aS)-8-amino-1-oxo-5-(2-phenylethyl)hexahydro-1H-pyrrolo[1,2-a][1,4]diazepin-2(3H)-yl]-N-[(3,4-dichlorophenyl)methyl]-4-methylpentanamide, POTASSIUM ION, Three-prime repair exonuclease 1, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Xu, J, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-01 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9AUC
 
 | | Human Amylin1 Receptor in Complex with Gs and human Calcitonin Gene-Related Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin gene-related peptide 1, ... | | Authors: | Cao, J, Belousoff, M.J, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2024-02-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM Structure of the Human Amylin 1 Receptor in Complex with CGRP and Gs Protein.
Biochemistry, 63, 2024
|
|
9ATT
 
 | | Crystal structure of MERS 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (R-enantiomer) | | Descriptor: | (1R,2S)-2-{[N-({[(2R)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2R)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-27 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9ATS
 
 | | Crystal structure of MERS 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (S-enantiomer) | | Descriptor: | (1S,2S)-2-{[N-({[(2S)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-27 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9ATJ
 
 | | Crystal structure of MERS 3CL protease in complex with a m-chlorobenzyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
9ATI
 
 | | Crystal structure of MERS 3CL protease in complex with a racemic bicyclo[2.2.1]heptenyl-methyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(3S)-1-{[(1R,2R,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(3S)-1-{[(1R,2R,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 2024
|
|
