1YB9
 
 | | Crystal structure of the A-DNA GCGTAT*CGC with a 2'-O-[2-(N,N-dimethylaminooxy)ethyl] Thymidine (T*) | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(2OT)P*AP*CP*GP*C)-3'), SPERMINE | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharan, M. | | Deposit date: | 2004-12-20 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Probing the Influence of Stereoelectronic Effects on the Biophysical Properties of Oligonucleotides: Comprehensive Analysis of the RNA Affinity, Nuclease Resistance, and Crystal Structure of Ten 2'-O-Ribonucleic Acid Modifications.
Biochemistry, 44, 2005
|
|
1JN5
 
 | | Structural basis for the recognition of a nucleoporin FG-repeat by the NTF2-like domain of TAP-p15 mRNA export factor | | Descriptor: | FG-repeat, TAP, p15 | | Authors: | Fribourg, S, Braun, I.C, Izaurralde, E, Conti, E. | | Deposit date: | 2001-07-23 | | Release date: | 2001-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor.
Mol.Cell, 8, 2001
|
|
1JKG
 
 | | Structural basis for the recognition of a nucleoporin FG-repeat by the NTF2-like domain of TAP-p15 mRNA nuclear export factor | | Descriptor: | TAP, p15 | | Authors: | Fribourg, S, Braun, I.C, Izaurralde, E, Conti, E. | | Deposit date: | 2001-07-12 | | Release date: | 2001-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor.
Mol.Cell, 8, 2001
|
|
1D8Y
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF DNA POLYMERASE I KLENOW FRAGMENT WITH DNA | | Descriptor: | D(T)19 OLIGOMER, DNA POLYMERASE I, SULFATE ION, ... | | Authors: | Teplova, M, Wallace, S.T, Tereshko, V, Minasov, G, Simons, A.M, Cook, P.D, Manoharan, M, Egli, M. | | Deposit date: | 1999-10-26 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural origins of the exonuclease resistance of a zwitterionic RNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2L9J
 
 | | hRSV M2-1 core domain structure | | Descriptor: | Matrix protein 2-1 | | Authors: | Dubosclard, V, Blondot, M, Bontems, F, Eleouet, J, Sizun, C. | | Deposit date: | 2011-02-12 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and functional analysis of the RNA- and viral phosphoprotein-binding domain of respiratory syncytial virus M2-1 protein.
Plos Pathog., 8, 2012
|
|
6RJ9
 
 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA20-AcrIIA6 monomeric assembly. | | Descriptor: | AcrIIA6, CRISPR-associated endonuclease Cas9 1, sgRNA, ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
1MSC
 
 | |
4ILR
 
 | |
6C54
 
 | | Ebola nucleoprotein nucleocapsid-like assembly and the asymmetric unit | | Descriptor: | Nucleoprotein | | Authors: | Su, Z, Wu, C, Pintilie, G.D, Chiu, W, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2018-01-13 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Electron Cryo-microscopy Structure of Ebola Virus Nucleoprotein Reveals a Mechanism for Nucleocapsid-like Assembly.
Cell, 172, 2018
|
|
4NIA
 
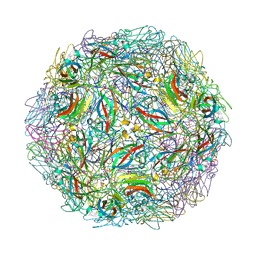 | | Satellite Tobacco Mosaic Virus Refined at room temperature to 1.8 A Resolution using NCS Restraints | | Descriptor: | Coat protein, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Satellite tobacco mosaic virus refined to 1.4 angstrom resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
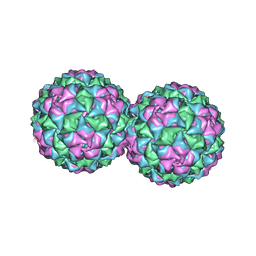
2VF9
 
 | |
6RJD
 
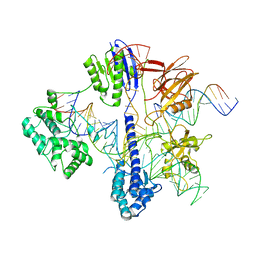 | | Cryo-EM structure of St1Cas9-sgRNA-tDNA59-ntPAM complex. | | Descriptor: | Streptococcus Thermophilus 1 Cas9, ntPAM, sgRNA (78-MER), ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
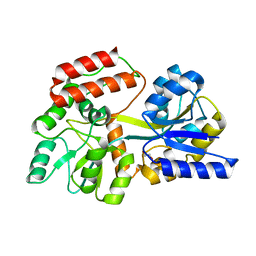
3HST
 
 | |
6RJG
 
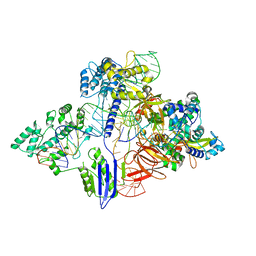 | | Cryo-EM structure of St1Cas9-sgRNA-AcrIIA6-tDNA59-ntPAM complex. | | Descriptor: | AcrIIA6, Cas 9, ntPAM, ... | | Authors: | Goulet, A, Chaves-Sanjuan, A, Cambillau, C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6.
Mol.Cell, 76, 2019
|
|
4D2G
 
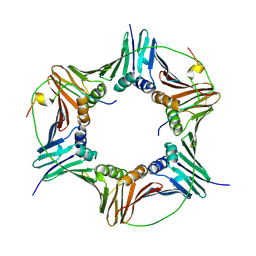 | | Crystal structure of human PCNA in complex with p15 peptide | | Descriptor: | P15, PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | DeBiasio, A, Ibanez, A, Mortuza, G, Molina, R, Cordeiro, T.N, Castillo, F, Villate, M, Merino, N, Lelli, M, Diercks, T, Luque, I, Bernardo, P, Montoya, G, Blanco, F.J. | | Deposit date: | 2014-05-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of P15(Paf)-PCNA Complex and Implications for Clamp Sliding During DNA Replication and Repair.
Nat.Commun., 6, 2015
|
|
2HT1
 
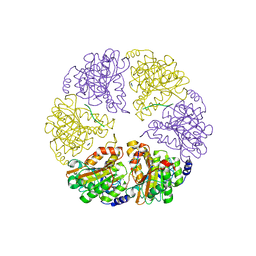 | |
1QGP
 
 | | NMR STRUCTURE OF THE Z-ALPHA DOMAIN OF ADAR1, 15 STRUCTURES | | Descriptor: | PROTEIN (DOUBLE STRANDED RNA ADENOSINE DEAMINASE) | | Authors: | Schade, M, Turner, C.J, Kuehne, R, Schmieder, P, Lowenhaupt, K, Herbert, A, Rich, A, Oschkinat, H. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3LP3
 
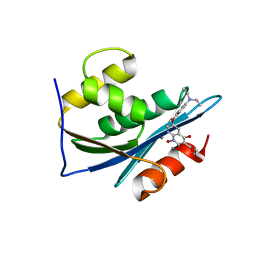 | | p15 HIV RNaseH domain with inhibitor MK3 | | Descriptor: | 3-[4-(diethylamino)phenoxy]-6-(ethoxycarbonyl)-5,8-dihydroxy-7-oxo-7,8-dihydro-1,8-naphthyridin-1-ium, MANGANESE (II) ION, p15 | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
1DT4
 
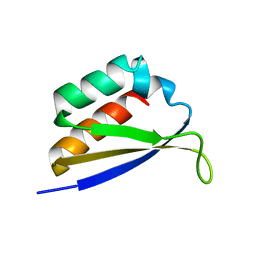 | | CRYSTAL STRUCTURE OF NOVA-1 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | NEURO-ONCOLOGICAL VENTRAL ANTIGEN 1 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L. | | Deposit date: | 2000-01-11 | | Release date: | 2000-02-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
2HB5
 
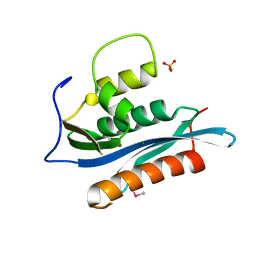 | | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain | | Descriptor: | MAGNESIUM ION, Reverse transcriptase/ribonuclease H, SULFATE ION | | Authors: | Lim, D, Gregorio, G.G, Bingman, C.A, Martinez-Hackert, E, Hendrickson, W.A, Goff, S.P. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain.
J.Virol., 80, 2006
|
|
4GEL
 
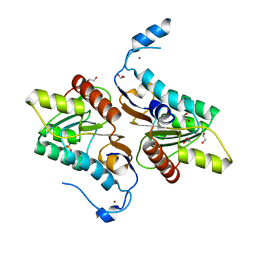 | | Crystal structure of Zucchini | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, PHOSPHATE ION, ... | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
3HHN
 
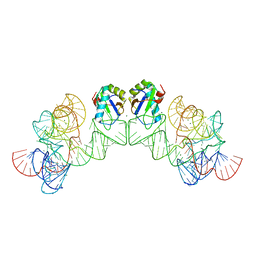 | | Crystal structure of class I ligase ribozyme self-ligation product, in complex with U1A RBD | | Descriptor: | Class I ligase ribozyme, self-ligation product, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Grant, R.A, Bagby, S.C, Bartel, D.P. | | Deposit date: | 2009-05-15 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.987 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
4FTS
 
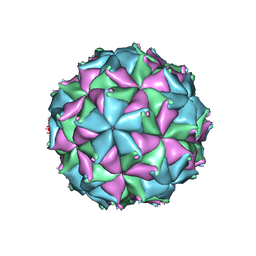 | | Crystal structure of the N363T mutant of the Flock House virus capsid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
3AGV
 
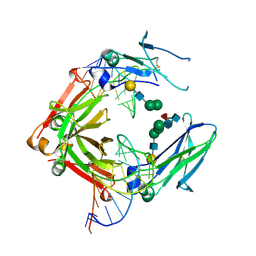 | | Crystal structure of a human IgG-aptamer complex | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*(UFT)P*GP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*GP*AP*AP*A*GP*GP*AP*AP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*A)-3', CALCIUM ION, Ig gamma-1 chain C region, ... | | Authors: | Nomura, Y, Sugiyama, S, Sakamoto, T, Miyakawa, S, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Nakamura, Y, Matsumura, H. | | Deposit date: | 2010-04-08 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational plasticity of RNA for target recognition as revealed by the 2.15 A crystal structure of a human IgG-aptamer complex
Nucleic Acids Res., 38, 2010
|
|
2R70
 
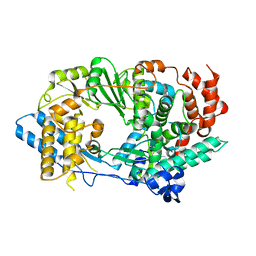 | | Crystal structure of infectious bursal disease virus VP1 polymerase, cocrystallized with an oligopeptide mimicking the VP3 C-terminus. | | Descriptor: | INFECTIOUS BURSAL VIRUS VP1 POLYMERASE | | Authors: | Garriga, D, Navarro, A, Querol-Audi, J, Abaitua, F, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-09-07 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activation mechanism of a noncanonical RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
