8UQT
 
 | | Crystal structure of the Tree Shrew p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53, SULFATE ION | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
9DLH
 
 | | donor substrate analog-bound AftB | | Descriptor: | 2-deoxy-2-fluoro-1-O-[(S)-hydroxy{[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]oxy}phosphoryl]-beta-D-arabinofuranose, Arabinosyltransferase AftB | | Authors: | Yaqi, L. | | Deposit date: | 2024-09-11 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
9DLF
 
 | | Arabinosyltransferase AftB in complex with Fab_B3 | | Descriptor: | Arabinosyltransferase AftB, Fab_B3 heavy chain, Fab_B3 light chain, ... | | Authors: | Liu, Y, Mancia, F. | | Deposit date: | 2024-09-10 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
8UQR
 
 | | Crystal structure of the human p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
8UQS
 
 | | Crystal structure of the Opossum p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53 (Fragment) | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
9DM7
 
 | | mannosyltransferase PimE in complex with Fab_E6 | | Descriptor: | Fab_E6 heavy chain, Fab_E6 light chain, Mannosyltransferase | | Authors: | Liu, Y. | | Deposit date: | 2024-09-12 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
9DM5
 
 | | Product-Bound mannosyltransferase PimE | | Descriptor: | (2S)-1-{[(S)-{[(1S,2R,3R,4S,5S,6R)-2-[(6-O-hexadecanoyl-beta-L-gulopyranosyl)oxy]-3,4,5-trihydroxy-6-{[beta-D-mannopyranosyl-(1->2)-alpha-D-mannopyranosyl-(1->6)-beta-D-mannopyranosyl-(1->6)-alpha-D-mannopyranosyl]oxy}cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}-3-(hexadecanoyloxy)propan-2-yl 10-methyloctadecanoate, MONO-TRANS, OCTA-CIS DECAPRENYL-PHOSPHATE, ... | | Authors: | Liu, Y. | | Deposit date: | 2024-09-12 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Mechanistic studies of mycobacterial glycolipid biosynthesis by the mannosyltransferase PimE.
Nat Commun, 16, 2025
|
|
6RS8
 
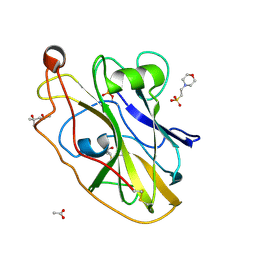 | | X-ray crystal structure of LsAA9B (transition metals soak) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
7N44
 
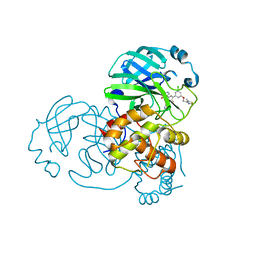 | | Crystal structure of the SARS-CoV-2 (2019-NCoV) main protease in complex with 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione (compound 13) | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Reilly, R.A, Zhang, C.H, Deshmukh, M.G, Ippolito, J.A, Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-06-03 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency.
Acs Med.Chem.Lett., 12, 2021
|
|
6EUE
 
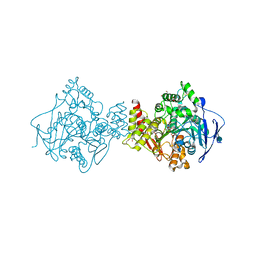 | | Rivastigmine analogue bound to Tc ACHE. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, PENTAETHYLENE GLYCOL | | Authors: | De la Mora, E, Brazzolotto, X, Dighe, S.N, Silman, I, Weik, M, Ross, B. | | Deposit date: | 2017-10-30 | | Release date: | 2018-11-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rivastigmine and metabolite analogues with putative Alzheimer's disease-modifying properties in a Caenorhabditis elegans model
Commun Chem, 2019
|
|
2CLY
 
 | | Subcomplex of the stator of bovine mitochondrial ATP synthase | | Descriptor: | ATP SYNTHASE B CHAIN, MITOCHONDRIAL, ATP SYNTHASE COUPLING FACTOR 6, ... | | Authors: | Kane Dickson, V, Silvester, J.A, Fearnley, I.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | On the Structure of the Stator of the Mitochondrial ATP Synthase.
Embo J., 25, 2006
|
|
5UV6
 
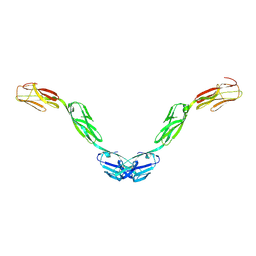 | | Crystal structure of human Opioid Binding Protein/Cell Adhesion Molecule Like (OPCML) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Opioid-binding protein/cell adhesion molecule, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Birtley, J.R, Stern, L.J, Gabra, H, Zanini, E. | | Deposit date: | 2017-02-19 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65002 Å) | | Cite: | Inactivating mutations and X-ray crystal structure of the tumor suppressor OPCML reveal cancer-associated functions.
Nat Commun, 10, 2019
|
|
6K7Y
 
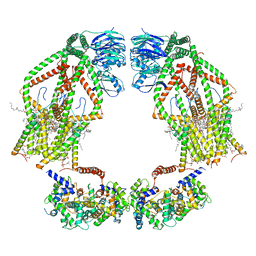 | | Intact human mitochondrial calcium uniporter complex with MICU1/MICU2 subunits | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhuo, W, Zhou, H, Yang, M. | | Deposit date: | 2019-06-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Protein Cell, 12, 2021
|
|
7NNH
 
 | |
6MID
 
 | |
1OMY
 
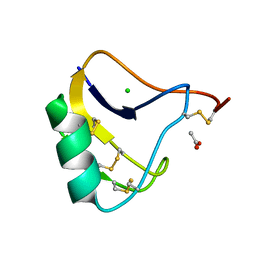 | | Crystal Structure of a Recombinant alpha-insect Toxin BmKaIT1 from the scorpion Buthus martensii Karsch | | Descriptor: | ACETIC ACID, Alpha-neurotoxin TX12, CHLORIDE ION | | Authors: | Huang, Y, Huang, Q, Chen, H, Tang, Y, Miyake, H, Kusunoki, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary crystallographic study of rBmKalphaIT1, a recombinant alpha-insect toxin from the scorpion Buthus martensii Karsch.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2NNE
 
 | | The Structural Identification of the Interaction Site and Functional State of RBP for its Membrane Receptor | | Descriptor: | CADMIUM ION, GLYCEROL, Major urinary protein 2 | | Authors: | Redondo, C, Bingham, R.J, Vouropoulou, M, Homans, S.W, Findlay, J.B. | | Deposit date: | 2006-10-24 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of the retinol-binding protein (RBP) interaction site and functional state of RBPs for the membrane receptor.
Faseb J., 22, 2008
|
|
6D3M
 
 | | FT_T dioxygenase with bound quizalofop | | Descriptor: | (2R)-2-{4-[(6-chloroquinoxalin-2-yl)oxy]phenoxy}propanoic acid, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
6CT2
 
 | | MYST histone acetyltransferase KAT6A/B in complex with WM-1119 | | Descriptor: | 3-fluoro-N'-[(2-fluorophenyl)sulfonyl]-5-(pyridin-2-yl)benzohydrazide, Histone acetyltransferase KAT8, MAGNESIUM ION, ... | | Authors: | Ren, B, Peat, T.S. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
6MQC
 
 | |
3NM8
 
 | | Crystal structure of Tyrosinase from Bacillus megaterium | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Tyrosinase, ... | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-06-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity
J.Mol.Biol., 405, 2011
|
|
5O2F
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed ampicillin - new refinement | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
7FCD
 
 | | Structure of the SARS-CoV-2 A372T spike glycoprotein (open) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Loss of Spike N370 glycosylation as an important evolutionary event for the enhanced infectivity of SARS-CoV-2.
Cell Res., 32, 2022
|
|
7FCE
 
 | | Structure of the SARS-CoV-2 A372T spike glycoprotein (closed) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Loss of Spike N370 glycosylation as an important evolutionary event for the enhanced infectivity of SARS-CoV-2.
Cell Res., 32, 2022
|
|
6KD1
 
 | |
