6LF4
 
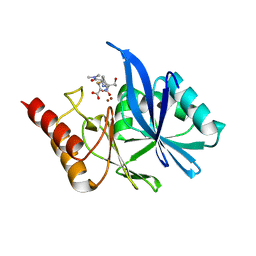 | | Crystal structure of VMB-1 bound to hydrolyzed meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, VMB-1 metallo-beta-lactamase, ZINC ION | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-11-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of VMB-1 bound to hydrolyzed meropenem
To Be Published
|
|
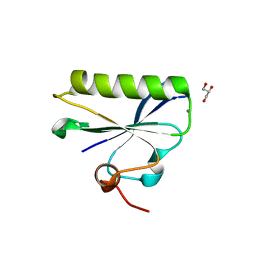
6LJE
 
 | |
6L9Z
 
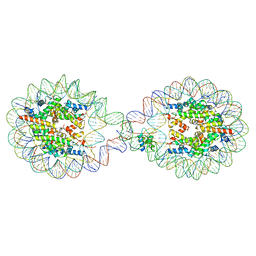 | | 338 bp di-nucleosome assembled with linker histone H1.X | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (338-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LBM
 
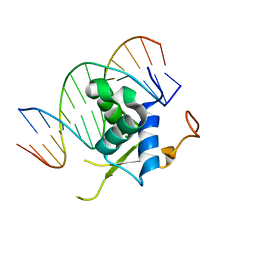 | |
6LBI
 
 | |
6LAB
 
 | | 169 bp nucleosome, harboring cohesive DNA termini, assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Bao, Q, Lee, P.L, Padavattan, S, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LA2
 
 | | 343 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | DNA (343-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
6LER
 
 | | 169 bp nucleosome harboring non-identical cohesive DNA termini. | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Sharma, D, Adhireksan, Z, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-26 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6L4J
 
 | |
6L44
 
 | |
6L4N
 
 | |
6L4I
 
 | |
4UX2
 
 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. MgF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
5I59
 
 | | Glutamate- and glycine-bound GluN1/GluN2A agonist binding domains with MPX 007 | | Descriptor: | 5-({[(3,4-difluorophenyl)sulfonyl]amino}methyl)-6-methyl-N-[(2-methyl-4H-1lambda~4~,3-thiazol-5-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
6LPW
 
 | |
6LPF
 
 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-10 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
6LNP
 
 | | Crystal structure of citrate Biosensor | | Descriptor: | CITRIC ACID, Fusion protein of Green fluorescent protein and Sensor histidine kinase CitA | | Authors: | Wen, Y, Campbell, R. | | Deposit date: | 2019-12-31 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | High-Performance Intensiometric Direct- and Inverse-Response Genetically Encoded Biosensors for Citrate.
Acs Cent.Sci., 6, 2020
|
|
6LLG
 
 | | Crystal Structure of Fagopyrum esculentum M UGT708C1 | | Descriptor: | BENZAMIDINE, SULFATE ION, UDP-glycosyltransferase 708C1 | | Authors: | Wang, X, Liu, M. | | Deposit date: | 2019-12-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of theC-Glycosyltransferase UGT708C1 from Buckwheat Provide Insights into the Mechanism ofC-Glycosylation.
Plant Cell, 32, 2020
|
|
6LNQ
 
 | | The co-crystal structure of SARS-CoV 3C Like Protease with aldehyde inhibitor M7 | | Descriptor: | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]-1H-indole-2-carboxamide, Severe Acute Respiratory Syndrome Coronavirus 3c Like Protease | | Authors: | Wang, H, Shang, L.Q. | | Deposit date: | 2020-01-01 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Comprehensive Insights into the Catalytic Mechanism of Middle East Respiratory Syndrome 3C-Like Protease and Severe Acute Respiratory Syndrome 3C-Like Protease.
Acs Catalysis, 10, 2020
|
|
4FYI
 
 | | Crystal structure of rcl with 6-cyclopentyl-AMP | | Descriptor: | Deoxyribonucleoside 5'-monophosphate N-glycosidase, N-cyclopentyladenosine 5'-(dihydrogen phosphate) | | Authors: | Labesse, G, Padilla, A, Kaminski, P.A. | | Deposit date: | 2012-07-04 | | Release date: | 2013-01-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of the oncoprotein Rcl bound to three nucleotide analogues.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VYV
 
 | | Crystal structure of subtilisin NAT at 1.36 | | Descriptor: | CALCIUM ION, GLYCEROL, Subtilisin NAT | | Authors: | Ushijima, H, Fuchita, N, Kajiwara, T, Motoshima, H, Ueno, G, Watanabe, K. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of subtilisin NAT at 1.36
TO BE PUBLISHED
|
|
4UQJ
 
 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | Descriptor: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
6LX4
 
 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LKT
 
 | | Crystal structure of the Fab fragment of murine monoclonal antibody KH-1 against Human herpesvirus 6B | | Descriptor: | antibody Fab Fragment L-chain, antibody Fab fragment H chain | | Authors: | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | Deposit date: | 2019-12-20 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
