1NUJ
 
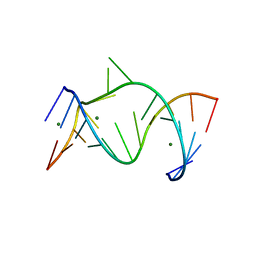 | | THE LEADZYME STRUCTURE BOUND TO MG(H20)6(II) AT 1.8 A RESOLUTION | | Descriptor: | 5'-R(*CP*GP*GP*AP*CP*CP*GP*AP*GP*CP*CP*AP*G)-3', 5'-R(*GP*CP*UP*GP*GP*GP*AP*GP*UP*CP*C)-3', MAGNESIUM ION | | Authors: | Wedekind, J.E, Mckay, D.B. | | Deposit date: | 2003-01-31 | | Release date: | 2003-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the leadzyme at 1.8 A resolution: metal ion binding and the implications
for catalytic mechanism and allo site ion regulation.
BIOCHEMISTRY, 42, 2003
|
|
1NW9
 
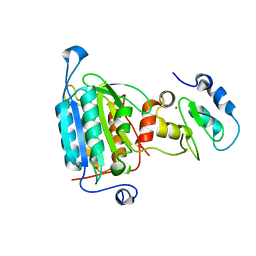 | | STRUCTURE OF CASPASE-9 IN AN INHIBITORY COMPLEX WITH XIAP-BIR3 | | Descriptor: | Baculoviral IAP repeat-containing protein 4, ZINC ION, caspase 9, ... | | Authors: | Shiozaki, E.N, Chai, J, Rigotti, D.J, Riedl, S.J, Li, P, Srinivasula, S.M, Alnemri, E.S, Fairman, R, Shi, Y. | | Deposit date: | 2003-02-05 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of XIAP-Mediated Inhibition of Caspase-9
Mol.Cell, 11, 2003
|
|
6UJB
 
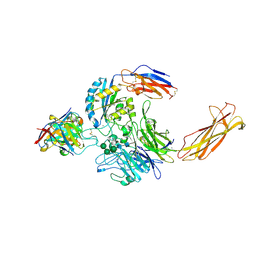 | | Integrin alpha-v beta-8 in complex with the Fabs C6D4 and 11D12v2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C6D4 heavy chain Fab, ... | | Authors: | Campbell, M.G, Cormier, A, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM Reveals Integrin-Mediated TGF-beta Activation without Release from Latent TGF-beta.
Cell, 180, 2020
|
|
6UJS
 
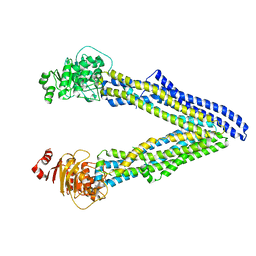 | | P-glycoprotein mutant-F728A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
1BCB
 
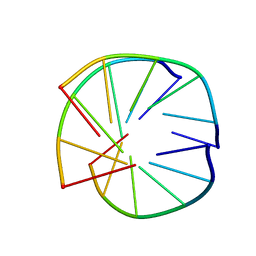 | | INTRAMOLECULAR TRIPLEX, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*T)-3') | | Authors: | Asensio, J.L, Brown, T, Lane, A.N. | | Deposit date: | 1998-04-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of intramolecular DNA triple helices containing adjacent and non-adjacent CG.C+ triplets.
Nucleic Acids Res., 26, 1998
|
|
6UJN
 
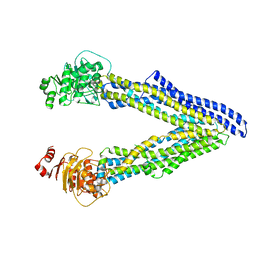 | | P-glycoprotein mutant-C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
2K9E
 
 | |
1FV1
 
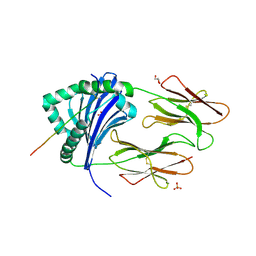 | | STRUCTURAL BASIS FOR THE BINDING OF AN IMMUNODOMINANT PEPTIDE FROM MYELIN BASIC PROTEIN IN DIFFERENT REGISTERS BY TWO HLA-DR2 ALLELES | | Descriptor: | GLYCEROL, MAJOR HISTOCOMPATIBILITY COMPLEX ALPHA CHAIN, MAJOR HISTOCOMPATIBILITY COMPLEX BETA CHAIN, ... | | Authors: | Li, H, Mariuzza, A.R, Li, Y, Martin, R. | | Deposit date: | 2000-09-18 | | Release date: | 2000-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.
J.Mol.Biol., 304, 2000
|
|
2K6D
 
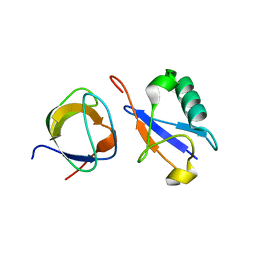 | | CIN85 Sh3-C domain in complex with ubiquitin | | Descriptor: | SH3 domain-containing kinase-binding protein 1, Ubiquitin | | Authors: | Forman-Kay, J, Bezsonova, I. | | Deposit date: | 2008-07-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interactions between the three CIN85 SH3 domains and ubiquitin: implications for CIN85 ubiquitination.
Biochemistry, 47, 2008
|
|
1HUI
 
 | | INSULIN MUTANT (B1, B10, B16, B27)GLU, DES-B30, NMR, 25 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Olsen, H.B, Ludvigsen, S, Kaarsholm, N.C. | | Deposit date: | 1996-03-29 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an engineered insulin monomer at neutral pH.
Biochemistry, 35, 1996
|
|
2KA9
 
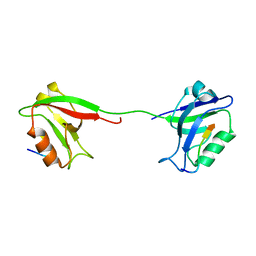 | | Solution structure of PSD-95 PDZ12 complexed with cypin peptide | | Descriptor: | Disks large homolog 4, cypin peptide | | Authors: | Wang, W.N, Weng, J.W, Zhang, X, Liu, M.L, Zhang, M.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Creating conformational entropy by increasing interdomain mobility in ligand binding regulation: a revisit to N-terminal tandem PDZ domains of PSD-95
J.Am.Chem.Soc., 131, 2009
|
|
6VBH
 
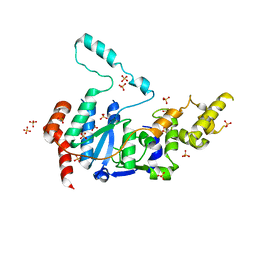 | | Human XPG endonuclease catalytic domain | | Descriptor: | DNA repair protein complementing XP-G cells,Flap endonuclease 1, SULFATE ION | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1B4O
 
 | |
6USZ
 
 | | Identification of the Clinical Development Candidate MRTX849, a Covalent KRASG12C Inhibitor for the Treatment of Cancer | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Vigers, G.P, Smith, D.J. | | Deposit date: | 2019-10-28 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of the Clinical Development CandidateMRTX849, a Covalent KRASG12CInhibitor for the Treatment of Cancer.
J.Med.Chem., 63, 2020
|
|
1BHL
 
 | | CACODYLATED CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-06-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1AQQ
 
 | | AG-SUBSTITUTED METALLOTHIONEIN FROM SACCHAROMYCES CEREVISIAE, NMR, 10 STRUCTURES | | Descriptor: | AG-METALLOTHIONEIN, SILVER ION | | Authors: | Peterson, C.W, Narula, S.S, Armitage, I.M. | | Deposit date: | 1997-07-31 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 3D solution structure of copper and silver-substituted yeast metallothioneins.
FEBS Lett., 379, 1996
|
|
2KTT
 
 | | Solution Structure of a Covalently Bound Pyrrolo[2,1-c][1,4]benzodiazepine-Benzimidazole Hybrid to a 10mer DNA Duplex | | Descriptor: | (11aS)-7-methoxy-8-(3-{4-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]phenoxy}propoxy)-1,2,3,10,11,11a-hexahydro-5H-pyrrolo[2,1-c][1,4]benzodiazepin-5-one, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Weingarth, M, Langel, W, Kamal, A, Kumar, P.P, Weisz, K. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a covalently bound pyrrolo[2,1-c][1,4]benzodiazepine-benzimidazole hybrid to a 10mer DNA duplex.
Biochemistry, 48, 2009
|
|
6VAL
 
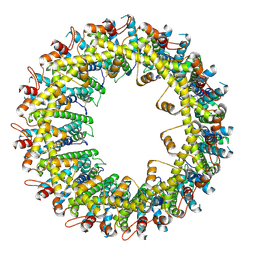 | |
7DTO
 
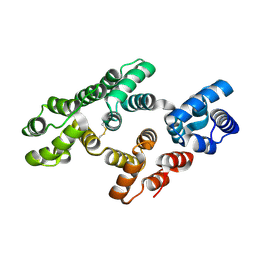 | |
1BZ7
 
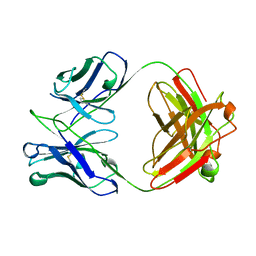 | | FAB FRAGMENT FROM MURINE ASCITES | | Descriptor: | PROTEIN (ANTIBODY R24 (HEAVY CHAIN)), PROTEIN (ANTIBODY R24 (LIGHT CHAIN)) | | Authors: | Kaminski, M.J, Mackenzie, C.R, Mooibroek, M.J, Dahms, T.E.S, Hirama, T, Houghton, A.N, Chapman, P.B, Evans, S.V. | | Deposit date: | 1998-11-06 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The role of homophilic binding in anti-tumor antibody R24 recognition of molecular surfaces. Demonstration of an intermolecular beta-sheet interaction between vh domains.
J.Biol.Chem., 274, 1999
|
|
2LQW
 
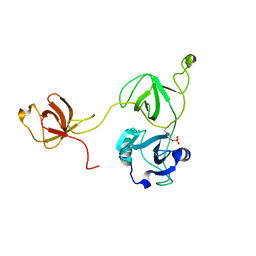 | |
1C1X
 
 | | L-PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND L-3-PHENYLLACTATE | | Descriptor: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, ISOPROPYL ALCOHOL, L-PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B. | | Deposit date: | 1999-07-22 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
Biochemistry, 39, 2000
|
|
1H0A
 
 | | Epsin ENTH bound to Ins(1,4,5)P3 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, EPSIN | | Authors: | Ford, M.G.J, McMahon, H.T, Evans, P.R. | | Deposit date: | 2002-06-12 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Curvature of Clathrin-Coated Pits Driven by Epsin
Nature, 419, 2002
|
|
7E3D
 
 | | Crystal structure of human acetylcholinesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Acetylcholinesterase | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2021-02-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
1BK4
 
 | | CRYSTAL STRUCTURE OF RABBIT LIVER FRUCTOSE-1,6-BISPHOSPHATASE AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEIN (FRUCTOSE-1,6-BISPHOSPHATASE), SULFATE ION | | Authors: | Ghosh, D, Weeks, C.M, Erman, M, Roszak, A.W, Kaiser, R, Jornvall, H. | | Deposit date: | 1998-07-14 | | Release date: | 1998-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of rabbit liver fructose 1,6-bisphosphatase at 2.3 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
