8GJ8
 
 | | RAD51C C-terminal domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAD51C | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
6RZX
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor FBSA | | Descriptor: | 1,1,2,2,3,3,4,4,4-nonakis(fluoranyl)butane-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
5A2H
 
 | | Crystal Structure of Arabidopsis thaliana Calmodulin-7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Kumar, S, Gourinath, S. | | Deposit date: | 2015-05-19 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Arabidopsis Thaliana Calmodulin7 and Insight Into its Mode of DNA Binding.
FEBS Lett., 590, 2016
|
|
8VTY
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with ciprofloxacin and protein Y at 2.60A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, 16S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 2024
|
|
8VTW
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-128 and protein Y at 2.35A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-cyclopropyl-7-(4-{4-[({[(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-14-(methoxyimino)-3a,7,9,11,13,15-hexamethyl-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl]oxy}carbonyl)amino]butyl}piperazin-1-yl)-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 2024
|
|
8BAR
 
 | |
2V1X
 
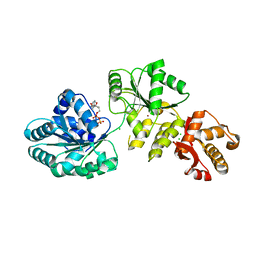 | | Crystal structure of human RECQ-like DNA helicase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT DNA HELICASE Q1, ... | | Authors: | Pike, A.C.W, Shrestha, B, Burgess-Brown, N, King, O, Ugochukwu, E, Watt, S, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Gileadi, O. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Human Recq1 Helicase Reveals a Putative Strand-Separation Pin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3BD4
 
 | |
4ODU
 
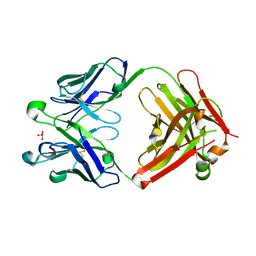 | |
1Z6A
 
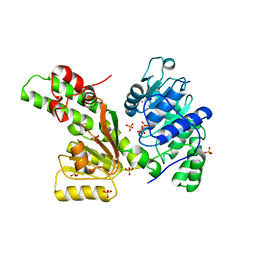 | | Sulfolobus solfataricus SWI2/SNF2 ATPase core domain | | Descriptor: | Helicase of the snf2/rad54 family, MERCURY (II) ION, PHOSPHATE ION | | Authors: | Duerr, H, Koerner, C, Mueller, M, Hickmann, V, Hopfner, K.P. | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA
Cell(Cambridge,Mass.), 121, 2005
|
|
4Q2C
 
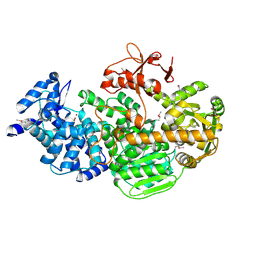 | | Crystal structure of CRISPR-associated protein | | Descriptor: | CRISPR-associated helicase Cas3, NICKEL (II) ION | | Authors: | Gong, B, Shin, M, Sun, J, van der Oost, J, Kim, J.-S. | | Deposit date: | 2014-04-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1N6Q
 
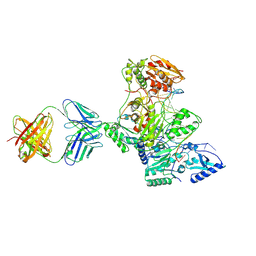 | | HIV-1 Reverse Transcriptase Crosslinked to pre-translocation AZTMP-terminated DNA (complex N) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3', 5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, I, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-11 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 Reverse Transcriptase with Pre- and Post-translocation AZTMP-terminated DNA
Embo J., 21, 2002
|
|
4ODW
 
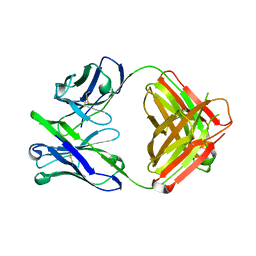 | | Unliganded Fab structure of lipid A-specific antibody A6 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, A6 Fab (IgG2b kappa) light chain, A6 Fab (IgG2b) heavy chain | | Authors: | Haji-Ghassemi, O, Evans, S.V. | | Deposit date: | 2014-01-10 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis for Antibody Recognition of Lipid A: INSIGHTS TO POLYSPECIFICITY TOWARD SINGLE-STRANDED DNA.
J.Biol.Chem., 290, 2015
|
|
7F9I
 
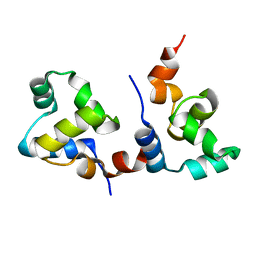 | | The apo-form structure of EnrR | | Descriptor: | EnrR repressor | | Authors: | Gan, J.H, Wang, Q.Y. | | Deposit date: | 2021-07-04 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding.
Nucleic Acids Res., 50, 2022
|
|
4Q2D
 
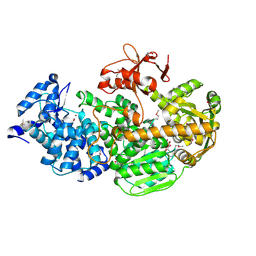 | | Crystal Structure of CRISPR-Associated protein in complex with 2'-Deoxyadenosine 5'-Triphosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CRISPR-associated helicase Cas3, MAGNESIUM ION, ... | | Authors: | Gong, B, Shin, M, Sun, J, van der Oost, J, Kim, J.-S. | | Deposit date: | 2014-04-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.771 Å) | | Cite: | Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1JMS
 
 | | Crystal Structure of the Catalytic Core of Murine Terminal Deoxynucleotidyl Transferase | | Descriptor: | MAGNESIUM ION, SODIUM ION, TERMINAL DEOXYNUCLEOTIDYLTRANSFERASE | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Sukumar, N, Jourdan, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-07-19 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
Embo J., 21, 2002
|
|
3IHU
 
 | |
1GJ2
 
 | | CO(III)-BLEOMYCIN-OOH BOUND TO AN OLIGONUCLEOTIDE CONTAINING A PHOSPHOGLYCOLATE LESION | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*CP*AP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3', ... | | Authors: | Hoehn, S.T, Junker, H.-D, Bunt, R.C, Turner, C.J, Stubbe, J. | | Deposit date: | 2000-11-01 | | Release date: | 2001-06-06 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Co(III)-bleomycin-OOH bound to a phosphoglycolate lesion containing oligonucleotide: implications for bleomycin-induced double-strand DNA cleavage.
Biochemistry, 40, 2001
|
|
7VP6
 
 | |
2WIZ
 
 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | ARCHAEAL HJC, HALF-JUNCTION | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
2WJ0
 
 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | ARCHAEAL HJC, COBALT HEXAMMINE(III), HALF-JUNCTION | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
8GXJ
 
 | |
8GXF
 
 | | Pseudomonas flexibilis GCN5 family acetyltransferase | | Descriptor: | COENZYME A, GCN5 family acetyltransferase | | Authors: | Song, Y.J, Bao, R. | | Deposit date: | 2022-09-20 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | The novel type II toxin-antitoxin PacTA modulates Pseudomonas aeruginosa iron homeostasis by obstructing the DNA-binding activity of Fur.
Nucleic Acids Res., 50, 2022
|
|
8GXK
 
 | | Pseudomonas jinjuensis N-acetyltransferase | | Descriptor: | COENZYME A, Protein N-acetyltransferase, RimJ/RimL family | | Authors: | Song, Y.J, Bao, R. | | Deposit date: | 2022-09-20 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The novel type II toxin-antitoxin PacTA modulates Pseudomonas aeruginosa iron homeostasis by obstructing the DNA-binding activity of Fur.
Nucleic Acids Res., 50, 2022
|
|
6ZPA
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and one catalytic Zn2+ ion | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.86000258 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
