3B2N
 
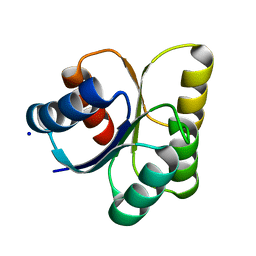 | | Crystal structure of DNA-binding response regulator, LuxR family, from Staphylococcus aureus | | Descriptor: | SODIUM ION, Uncharacterized protein Q99UF4 | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-18 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of DNA-binding response regulator, LuxR family, from Staphylococcus aureus.
To be Published
|
|
1MUS
 
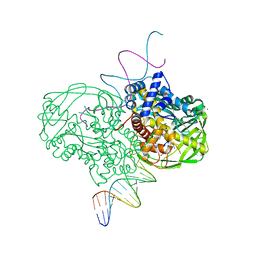 | | crystal structure of Tn5 transposase complexed with resolved outside end DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA non-transferred strand, DNA transferred strand, ... | | Authors: | Holden, H.M, Thoden, J.B, Steiniger-White, M, Reznikoff, W.S, Lovell, S, Rayment, I. | | Deposit date: | 2002-09-24 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure/function insights into Tn5 transposition.
Curr.Opin.Struct.Biol., 14, 2004
|
|
1Z19
 
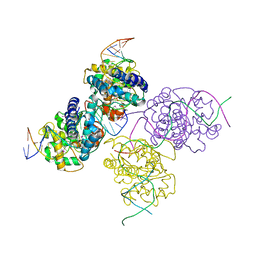 | | Crystal structure of a lambda integrase(75-356) dimer bound to a COC' core site | | Descriptor: | 33-MER, 5'-D(*CP*TP*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', 5'-D(P*TP*TP*TP*AP*TP*AP*CP*TP*AP*AP*GP*TP*TP*GP*GP*CP*AP*TP*TP*A)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
3VKX
 
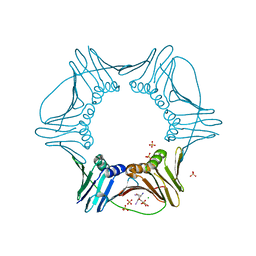 | | Structure of PCNA | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, CHLORIDE ION, Proliferating cell nuclear antigen, ... | | Authors: | Hashimoto, H, Hishiki, A, Shimizu, T, Sato, M, Punchihewa, C, Connelly, M, Actis, M, Waddell, B, Pagala, V, Fujii, N. | | Deposit date: | 2011-11-26 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication
J.Biol.Chem., 287, 2012
|
|
2B0E
 
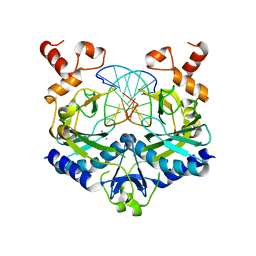 | | EcoRV Restriction Endonuclease/GAAUTC/Ca2+ | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*AP*(DU)P*TP*CP*TP*T)-3', CALCIUM ION, Type II restriction enzyme EcoRV | | Authors: | Hiller, D.A, Rodriguez, A.M, Perona, J.J. | | Deposit date: | 2005-09-13 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-cognate Enzyme-DNA Complex: Structural and Kinetic Analysis of EcoRV Endonuclease Bound to the EcoRI Recognition Site GAATTC
J.Mol.Biol., 354, 2005
|
|
4ENJ
 
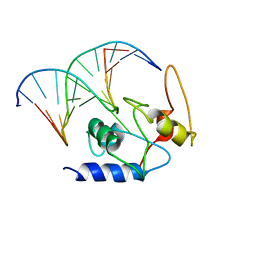 | |
3WGW
 
 | | Structure of PCNA bound to a small molecule inhibitor | | Descriptor: | 4-{4-[(2S)-2-amino-3-hydroxypropyl]-2,6-diiodophenoxy}phenol, Proliferating cell nuclear antigen, SULFATE ION | | Authors: | Hashimoto, H. | | Deposit date: | 2013-08-12 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A small molecule inhibitor of monoubiquitinated Proliferating Cell Nuclear Antigen (PCNA) inhibits repair of interstrand DNA cross-link, enhances DNA double strand break, and sensitizes cancer cells to cisplatin.
J.Biol.Chem., 289, 2014
|
|
1T05
 
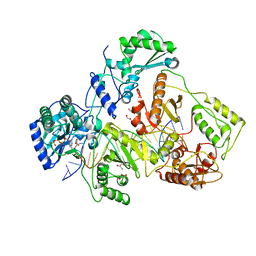 | | HIV-1 reverse transcriptase crosslinked to template-primer with tenofovir-diphosphate bound as the incoming nucleotide substrate | | Descriptor: | GLYCEROL, MAGNESIUM ION, POL polyprotein, ... | | Authors: | Tuske, S, Sarafianos, S.G, Ding, J, Arnold, E. | | Deposit date: | 2004-04-07 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RRR
 
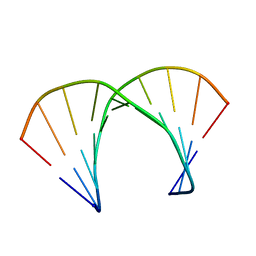 | | RNA DUPLEX CONTAINING A PURINE-RICH STRAND, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3'), RNA (5'-R(*GP*CP*UP*UP*CP*UP*CP*UP*UP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
2LF7
 
 | |
2LF8
 
 | |
6OJ0
 
 | | Cryo-EM reconstruction of Sulfolobus polyhedral virus 1 (SPV1) | | Descriptor: | Structural protein VP4, Uncharacterized protein | | Authors: | Wang, F, Liu, Y, Conway, J.F, Krupovic, M, Prangishvili, D, Egelman, E.H. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A packing for A-form DNA in an icosahedral virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1G3V
 
 | | CRYSTAL STRUCTURE OF NICKEL-D[CGTGTACACG]2 | | Descriptor: | 5'-D(*CP*GP*TP*GP*TP*AP*CP*AP*CP*G)-3', NICKEL (II) ION | | Authors: | Subirana, J.A, Abrescia, N.G.A. | | Deposit date: | 2000-10-25 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nickel-guanine interactions in DNA: crystal structure of nickel-d[CGTGTACACG]2.
J.Biol.Inorg.Chem., 7, 2002
|
|
5G4Q
 
 | | H.pylori Beta clamp in complex with 5-chloroisatin | | Descriptor: | 5-chloro-1H-indole-2,3-dione, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-16 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening of E. coli beta-clamp Inhibitors Revealed that Few Inhibit Helicobacter pylori More Effectively: Structural and Functional Characterization.
Antibiotics (Basel), 7, 2018
|
|
7OMU
 
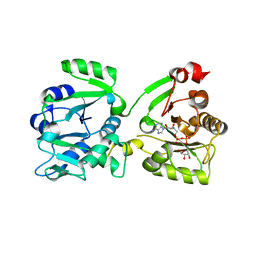 | | Thermosipho africanus DarTG in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Macro domain-containing protein | | Authors: | Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
8F27
 
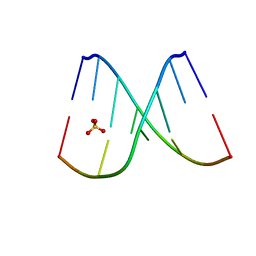 | | Mirror-image DNA containing 2'-OMe-L-uridine residue | | Descriptor: | Mirror-image DNA (5'-D(*(0DG)P*(0MU)P*(0DG)P*(0DT)P*(0DA)P*(0DC)P*(0DA)P*(0DC))-3'), SULFATE ION | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2022-11-07 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Derivatization of Mirror-Image l-Nucleic Acids with 2'-OMe Modification for Thermal and Structural Stabilization.
Chembiochem, 24, 2023
|
|
8KA4
 
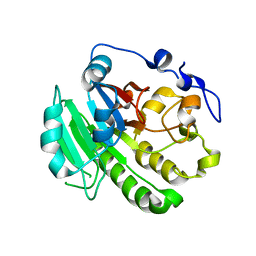 | |
6OZE
 
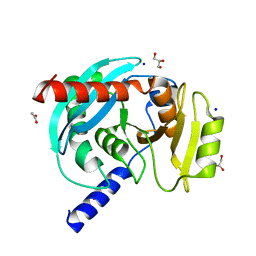 | |
4IC1
 
 | | Crystal structure of SSO0001 | | Descriptor: | IRON/SULFUR CLUSTER, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Lemak, S, Beloglazova, N, Flick, R, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-09 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus.
J.Am.Chem.Soc., 135, 2013
|
|
8HEY
 
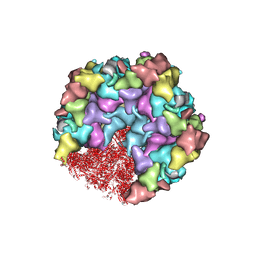 | | One CVSC-binding penton vertex in HCMV B-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HEX
 
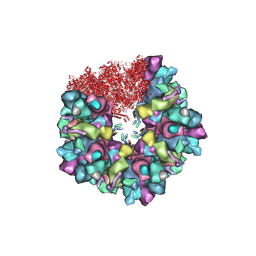 | | C5 portal vertex in HCMV B-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
6P81
 
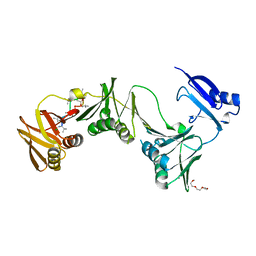 | |
6C66
 
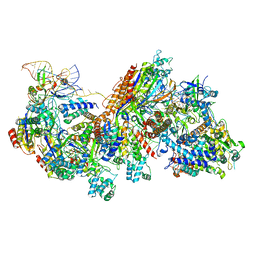 | | CRISPR RNA-guided surveillance complex, pre-nicking | | Descriptor: | CRISPR-associated helicase, Cas3 family, CRISPR-associated protein, ... | | Authors: | Xiao, Y, Luo, M, Liao, M, Ke, A. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure basis for RNA-guided DNA degradation by Cascade and Cas3.
Science, 361, 2018
|
|
5DKL
 
 | |
8OE3
 
 | |
