6ZEG
 
 | | Structure of PP1-IRSp53 chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZFX
 
 | | hSARM1 GraFix-ed | | Descriptor: | (~{E})-4-methylnon-4-enedial, NAD(+) hydrolase SARM1 | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-18 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
7OF2
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with GTPBP6. | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
2XI4
 
 | | Torpedo californica Acetylcholinesterase in Complex with Aflatoxin B1 (Orthorhombic Space Group) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, AFLATOXIN B1, ... | | Authors: | Sanson, B, Colletier, J.P, Xu, Y, Lang, P.T, Jiang, H, Silman, I, Sussman, J.L, Weik, M. | | Deposit date: | 2010-06-28 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Backdoor Opening Mechanism in Acetylcholinesterase Based on X-Ray Crystallography and Molecular Dynamics Simulations.
Protein Sci., 20, 2011
|
|
1MFQ
 
 | | Crystal Structure Analysis of a Ternary S-Domain Complex of Human Signal Recognition Particle | | Descriptor: | 7S RNA of human SRP, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kuglstatter, A, Oubridge, C, Nagai, K. | | Deposit date: | 2002-08-13 | | Release date: | 2002-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Induced structural changes of 7SL RNA during the assembly of human
signal recognition particle
Nat.Struct.Biol., 9, 2002
|
|
2XBG
 
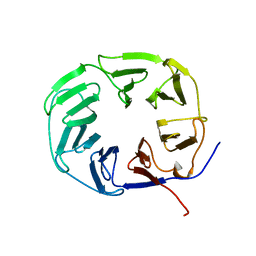 | |
1GR0
 
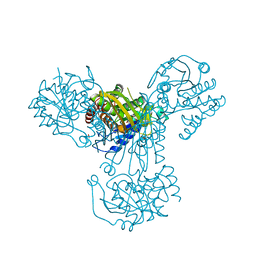 | | myo-inositol 1-phosphate synthase from Mycobacterium tuberculosis in complex with NAD and zinc. | | Descriptor: | CACODYLATE ION, INOSITOL-3-PHOSPHATE SYNTHASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Norman, R.A, Murray-Rust, J, McDonald, N.Q, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-10 | | Release date: | 2002-03-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Inositol 1-Phosphate Synthase from Mycobacterium Tuberculosis, a Key Enzyme in Phosphatidylinositol Synthesis
Structure, 10, 2002
|
|
1I8K
 
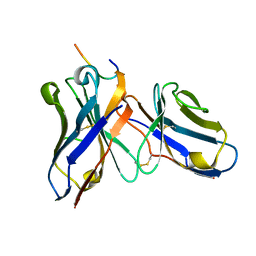 | | CRYSTAL STRUCTURE OF DSFV MR1 IN COMPLEX WITH THE PEPTIDE ANTIGEN OF THE MUTANT EPIDERMAL GROWTH FACTOR RECEPTOR, EGFRVIII, AT LIQUID NITROGEN TEMPERATURE | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR ANTIBODY MR1SCFV HEAVY CHAIN, EPIDERMAL GROWTH FACTOR RECEPTOR ANTIBODY MR1SCFV LIGHT CHAIN, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Landry, R.C, Klimowicz, A.C, Lavictoire, S.J, Borisova, S, Kottachchi, D.T, Lorimer, I.A, Evans, S.V. | | Deposit date: | 2001-03-14 | | Release date: | 2002-03-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antibody recognition of a conformational epitope in a peptide antigen: Fv-peptide complex of an antibody fragment specific for the mutant EGF receptor, EGFRvIII.
J.Mol.Biol., 308, 2001
|
|
1I39
 
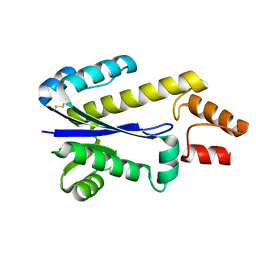 | | RNASE HII FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | RIBONUCLEASE HII | | Authors: | Chapados, B.R, Chai, Q, Hosfield, D.J, Qiu, J, Shen, B, Tainer, J.A. | | Deposit date: | 2001-02-13 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication.
J.Mol.Biol., 307, 2001
|
|
6Z01
 
 | | DNA Topoisomerase | | Descriptor: | CHLORIDE ION, DNA topoisomerase I | | Authors: | Takahashi, T.S, Gadelle, D, Forterre, P, Mayer, C, Petrella, S. | | Deposit date: | 2020-05-07 | | Release date: | 2021-11-17 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states.
Nat Commun, 13, 2022
|
|
6Z03
 
 | | DNA Topoisomerase | | Descriptor: | DNA topoisomerase I | | Authors: | Takahashi, T.S, Gadelle, D, Forterre, P, Mayer, C, Petrella, S. | | Deposit date: | 2020-05-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states.
Nat Commun, 13, 2022
|
|
1L9H
 
 | | Crystal structure of bovine rhodopsin at 2.6 angstroms RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Okada, T, Fujiyoshi, Y, Silow, M, Navarro, J, Landau, E.M, Shichida, Y. | | Deposit date: | 2002-03-23 | | Release date: | 2002-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1D62
 
 | |
7OYL
 
 | | Phosphoglucose isomerase of Aspergillus fumigatus in complexed with Glucose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raimi, O.G, Yan, K, Fang, W, van Aalten, D.M.F. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Phosphoglucose Isomerase Is Important for Aspergillus fumigatus Cell Wall Biogenesis.
Mbio, 13, 2022
|
|
2WWY
 
 | | Structure of human RECQ-like helicase in complex with a DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*DA DG DC DG DT DC DG DA DG DA DT DC DCP)-3', ATP-DEPENDENT DNA HELICASE Q1, ... | | Authors: | Pike, A.C.W, Zhang, Y, Schnecke, C, Chaikuad, A, Krojer, T, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recq1 Helicase-Driven DNA Unwinding, Annealing, and Branch Migration: Insights from DNA Complex Structures
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1LKT
 
 | | CRYSTAL STRUCTURE OF THE HEAD-BINDING DOMAIN OF PHAGE P22 TAILSPIKE PROTEIN | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage.
J.Mol.Biol., 267, 1997
|
|
1LJZ
 
 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | Descriptor: | Acetylcholine receptor protein, alpha-Bungarotoxin | | Authors: | Samson, A.O, Scherf, T, Eisenstein, M, Chill, J.H, Anglister, J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-07-17 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1I9X
 
 | |
1D5F
 
 | | STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY | | Descriptor: | E6AP HECT CATALYTIC DOMAIN, E3 LIGASE | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-07 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
1LD7
 
 | | Co-crystal structure of Human Farnesyltransferase with farnesyldiphosphate and inhibitor compound 66 | | Descriptor: | (20S)-19,20,22,23-TETRAHYDRO-19-OXO-5H,21H-18,20-ETHANO-12,14-ETHENO-6,10-METHENOBENZ[D]IMIDAZO[4,3-L][1,6,9,13]OXATRIA ZACYCLONOADECOSINE-9-CARBONITRILE, FARNESYL DIPHOSPHATE, ZINC ION, ... | | Authors: | Taylor, J.S, Terry, K.L, Beese, L.S. | | Deposit date: | 2002-04-08 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-Aminopyrrolidinone farnesyltransferase inhibitors: design of macrocyclic compounds with improved pharmacokinetics and excellent cell potency.
J.Med.Chem., 45, 2002
|
|
1LN6
 
 | | STRUCTURE OF BOVINE RHODOPSIN (Metarhodopsin II) | | Descriptor: | RETINAL, RHODOPSIN | | Authors: | Choi, G, Landin, J, Galan, J.F, Birge, R.R, Albert, A.D, Yeagle, P.L. | | Deposit date: | 2002-05-03 | | Release date: | 2002-07-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural studies of metarhodopsin II, the activated form of the G-protein coupled receptor, rhodopsin.
Biochemistry, 41, 2002
|
|
2Y2U
 
 | | Nonaged form of Mouse Acetylcholinesterase inhibited by VX-Update | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ACETYLCHOLINESTERASE, ... | | Authors: | Akfur, C, Artursson, E, Ekstrom, F. | | Deposit date: | 2010-12-16 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Methylphosphonate Adducts of Acetylcholinesterase Investigated by Time Correlated Single Photon Counting and X-Ray Crystallography
To be Published
|
|
1CEK
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE MEMBRANE-EMBEDDED M2 CHANNEL-LINING SEGMENT FROM THE NICOTINIC ACETYLCHOLINE RECEPTOR BY SOLID-STATE NMR SPECTROSCOPY | | Descriptor: | PROTEIN (ACETYLCHOLINE RECEPTOR M2) | | Authors: | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 1999-03-09 | | Release date: | 1999-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLID-STATE NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
6XDR
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 27 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-11 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XD0
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to NITD-434 | | Descriptor: | 2-[({2-[(2,6-dichlorophenyl)amino]phenyl}acetyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Arora, R, Benson, T.E, Liew, C.W, Lescar, J. | | Deposit date: | 2020-06-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Two RNA Tunnel Inhibitors Bind in Highly Conserved Sites in Dengue Virus NS5 Polymerase: Structural and Functional Studies.
J.Virol., 94, 2020
|
|
