2IO2
 
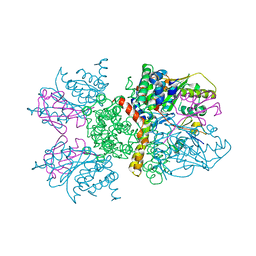 | | Crystal structure of human Senp2 in complex with RanGAP1-SUMO-1 | | Descriptor: | Ran GTPase-activating protein 1, Sentrin-specific protease 2, Small ubiquitin-related modifier 1 | | Authors: | Reverter, D, Lima, C.D. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates.
Nat.Struct.Mol.Biol., 13, 2006
|
|
8PPK
 
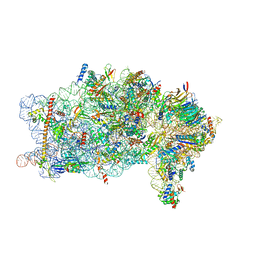 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
8PPL
 
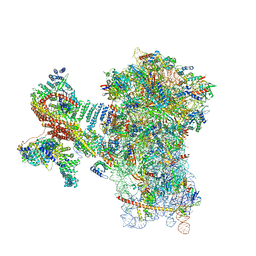 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
2HXY
 
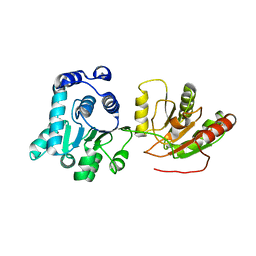 | | Crystal structure of human apo-eIF4AIII | | Descriptor: | Probable ATP-dependent RNA helicase DDX48 | | Authors: | Johansen, J.S, Andersen, G.R. | | Deposit date: | 2006-08-04 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA.
Science, 313, 2006
|
|
8ONS
 
 | |
2CQG
 
 | | Solution structure of the RNA binding domain of TAR DNA-binding protein-43 | | Descriptor: | TAR DNA-binding protein-43 | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of TAR DNA-binding protein-43
To be Published
|
|
2CS2
 
 | |
8P4F
 
 | | Structural insights into human co-transcriptional capping - structure 6 | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (38-MER), ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8OOA
 
 | | CryoEM Structure INO80core Hexasome complex Hexasome refinement state1 | | Descriptor: | DNA Strand 2, DNA strand 1, Histone H2A, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8PKI
 
 | | Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B type 1-C/E/G, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OJ0
 
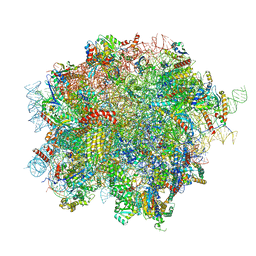 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OTS
 
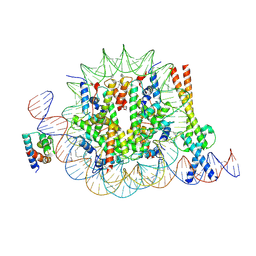 | | OCT4 and MYC-MAX co-bound to a nucleosome | | Descriptor: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8PKJ
 
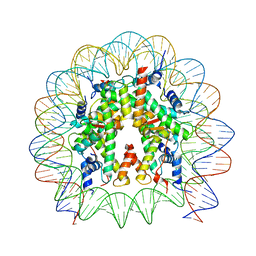 | | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2G50
 
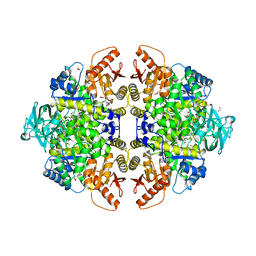 | | The location of the allosteric amino acid binding site of muscle pyruvate kinase. | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ALANINE, ... | | Authors: | Holyoak, T, Williams, R, Fenton, A.W. | | Deposit date: | 2006-02-22 | | Release date: | 2006-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differentiating a Ligand's Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase.
Biochemistry, 45, 2006
|
|
2G4D
 
 | | Crystal structure of human SENP1 mutant (C603S) in complex with SUMO-1 | | Descriptor: | SENP1 protein, Small ubiquitin-related modifier 1 | | Authors: | Xu, Z, Chau, S.F, Lam, K.H, Au, S.W.N. | | Deposit date: | 2006-02-22 | | Release date: | 2006-10-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the SENP1 mutant C603S-SUMO complex reveals the hydrolytic mechanism of SUMO-specific protease
Biochem.J., 398, 2006
|
|
2ED2
 
 | | Solution Structure of RSGI RUH-069, a GTF2I domain in human cDNA | | Descriptor: | General transcription factor II-I | | Authors: | Doi-Katayama, Y, Hirota, H, Izumi, K, Hayashi, F, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-14 | | Release date: | 2007-08-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-069, a GTF2I domain in human cDNA
To be published
|
|
2E42
 
 | | Crystal structure of C/EBPbeta Bzip homodimer V285A mutant bound to A High Affinity DNA fragment | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(P*DAP*DAP*DTP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DCP*DCP*DT)-3'), DNA (5'-D(P*DTP*DAP*DGP*DGP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DT)-3') | | Authors: | Tahirov, T.H, Inoue-Bungo, T, Sato, K, Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Flexible Base Recognition by C/Ebpbeta
To be Published
|
|
2E43
 
 | | Crystal structure of C/EBPbeta Bzip homodimer K269A mutant bound to A High Affinity DNA fragment | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(P*DAP*DAP*DTP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DCP*DCP*DT)-3'), DNA (5'-D(P*DTP*DAP*DGP*DGP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DT)-3') | | Authors: | Tahirov, T.H, Inoue-Bungo, T, Sato, K, Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Flexible Base Recognition by C/Ebpbeta
To be Published
|
|
2CUF
 
 | | Solution structure of the homeobox domain of the human hypothetical protein FLJ21616 | | Descriptor: | FLJ21616 protein | | Authors: | Ohnishi, S, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homeobox domain of the human hypothetical protein FLJ21616
To be Published
|
|
2CV5
 
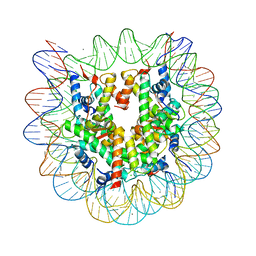 | | Crystal structure of human nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.a, ... | | Authors: | Tsunaka, Y, Kajimura, N, Tate, S, Morikawa, K. | | Deposit date: | 2005-05-31 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle
Nucleic Acids Res., 33, 2005
|
|
2KRR
 
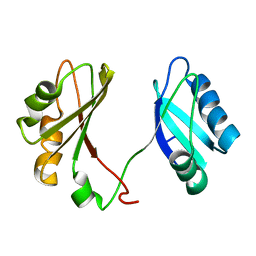 | | Solution structure of the RBD1,2 domains from human nucleolin | | Descriptor: | Nucleolin | | Authors: | Arumugam, N, Miller, C, Maliekal, J, Bates, P.J, Trent, J.O, Lane, A.N. | | Deposit date: | 2009-12-22 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RBD1,2 domains from human nucleolin.
J.Biomol.Nmr, 47, 2010
|
|
2D9B
 
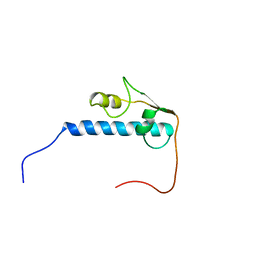 | | Solution Structure of RSGI RUH-052, a GTF2I domain in human cDNA | | Descriptor: | General transcription factor II-I | | Authors: | Doi-Katayama, Y, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-09 | | Release date: | 2006-06-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-052, a GTF2I domain in human cDNA
To be Published
|
|
2DO0
 
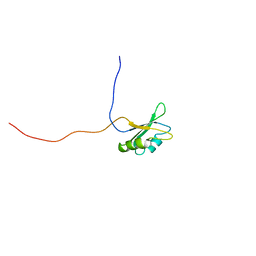 | | Solution structure of the RNA binding domain of heterogeneous nuclear ribonucleoprotein M | | Descriptor: | Heterogeneous nuclear ribonucleoprotein M | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-27 | | Release date: | 2006-10-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of heterogeneous nuclear ribonucleoprotein M
To be Published
|
|
2F9D
 
 | | 2.5 angstrom resolution structure of the spliceosomal protein p14 bound to region of SF3b155 | | Descriptor: | Pre-mRNA branch site protein p14, Splicing factor 3B subunit 1 | | Authors: | Schellenberg, M.J, Edwards, R.A, Ritchie, D.B, Glover, J.N.M, Macmillan, A.M. | | Deposit date: | 2005-12-05 | | Release date: | 2006-01-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a core spliceosomal protein interface
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
