5C5P
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | Descriptor: | (3R)-3-(1-hydroxy-2-methylpropan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2015-06-21 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
8ACL
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GC-14 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
5GZT
 
 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | Chitinase, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
5G2V
 
 | | Structure of BT4656 in complex with its substrate D-Glucosamine-2-N, 6-O-disulfate. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, N-ACETYLGLUCOSAMINE-6-SULFATASE, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8ACD
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GA-17S | | Descriptor: | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
5GZU
 
 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | Chitinase, PHOSPHATE ION | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
5H0I
 
 | |
5GZV
 
 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, PHOSPHATE ION | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
8G6Z
 
 | | JAK2 crystal structure in complex with Compound 13 | | Descriptor: | (3R)-3-cyclopentyl-3-[(4M)-4-{5-methyl-2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-1H-pyrazol-1-yl]propanenitrile, 1,2-ETHANEDIOL, Tyrosine-protein kinase JAK2 | | Authors: | Miller, S.T, Ellis, D.A. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Eyes on Topical Ocular Disposition: The Considered Design of a Lead Janus Kinase (JAK) Inhibitor That Utilizes a Unique Azetidin-3-Amino Bridging Scaffold to Attenuate Off-Target Kinase Activity, While Driving Potency and Aqueous Solubility.
J.Med.Chem., 66, 2023
|
|
8G8O
 
 | | The crystal structure of JAK2 in complex with Compound 31 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Tyrosine-protein kinase JAK2, ... | | Authors: | Miller, S.T, Ellis, D.A. | | Deposit date: | 2023-02-18 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Eyes on Topical Ocular Disposition: The Considered Design of a Lead Janus Kinase (JAK) Inhibitor That Utilizes a Unique Azetidin-3-Amino Bridging Scaffold to Attenuate Off-Target Kinase Activity, While Driving Potency and Aqueous Solubility.
J.Med.Chem., 66, 2023
|
|
5CGJ
 
 | | Crystal structure of murine Keap1 in complex with RA839, a non-covalent small-molecule binder to Keap1 and selective activator of Nrf2 signalling. | | Descriptor: | (3S)-1-(4-{[(2,3,5,6-tetramethylphenyl)sulfonyl]amino}naphthalen-1-yl)pyrrolidine-3-carboxylic acid, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Schimanski-Breves, S, Loenze, P, Engel, C.K. | | Deposit date: | 2015-07-09 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Characterization of RA839, a Noncovalent Small Molecule Binder to Keap1 and Selective Activator of Nrf2 Signaling.
J.Biol.Chem., 290, 2015
|
|
8G8X
 
 | | X-ray co-crystal structure of compound 27 in with complex JAK2 | | Descriptor: | 3-cyclopropyl-1-{5-methyl-2-[(3-methyl-1,2-thiazol-5-yl)amino]pyrimidin-4-yl}azetidin-3-ol, Tyrosine-protein kinase JAK2 | | Authors: | Miller, S.T, Ellis, D.A. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Eyes on Topical Ocular Disposition: The Considered Design of a Lead Janus Kinase (JAK) Inhibitor That Utilizes a Unique Azetidin-3-Amino Bridging Scaffold to Attenuate Off-Target Kinase Activity, While Driving Potency and Aqueous Solubility.
J.Med.Chem., 66, 2023
|
|
8GJS
 
 | | Stapled Peptide ALRN-6924 Bound to MDMX | | Descriptor: | ACE-LEU-THR-PHE-ALA-GLU-TYR-TRP-ALA-GLN-LEU-DAL-ALA-ALA-ALA-ALA-ALA-DAL, Protein Mdm4 | | Authors: | Graves, B.J, Janson, C, Lukacs, C. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Sulanemadlin (ALRN-6924), the First Cell-Permeating, Stabilized alpha-Helical Peptide in Clinical Development.
J.Med.Chem., 66, 2023
|
|
5CTT
 
 | |
2DYD
 
 | |
8JNP
 
 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with the substrate ikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450 CftA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, P, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5AL3
 
 | | Crystal structure of TNKS2 in complex with 2-(2,4-dichlorophenyl)-1- methyl-1H,2H,3H,4H-pyrido(2,3-d)pyrimidin-4-one | | Descriptor: | (2R)-2-(2,4-dichlorophenyl)-1-methyl-2,3-dihydropyrido[2,3-d]pyrimidin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Nkizinkiko, Y, Lehtio, L. | | Deposit date: | 2015-03-06 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Potent and Selective Nonplanar Tankyrase Inhibiting Nicotinamide Mimics.
Bioorg.Med.Chem., 23, 2015
|
|
8JNQ
 
 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with a substrate compound c | | Descriptor: | (1Z,3E,5E,7S,8R,10S,11R,13R,15R,16E,18E,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazatetracyclo[23.2.1.09,13.08,15]octacosa-1(2),3,5,16,18-pentaene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Jiang, P, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5AJA
 
 | | Crystal structure of mandrill SAMHD1 (amino acid residues 1-114) bound to Vpx isolated from mandrill and human DCAF1 (amino acid residues 1058-1396) | | Descriptor: | PROTEIN VPRBP, SAM DOMAIN AND HD DOMAIN-CONTAINING PROTEIN, VPX PROTEIN, ... | | Authors: | Schwefel, D, Boucherit, V.C, Christodoulou, E, Walker, P.A, Stoye, J.P, Bishop, K.N, Taylor, I.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Molecular Determinants for Recognition of Divergent Samhd1 Proteins by the Lentiviral Accessory Protein Vpx.
Cell Host Microbe., 17, 2015
|
|
8G2D
 
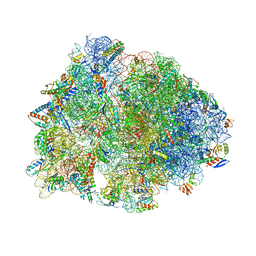 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with tylosin, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
5A51
 
 | |
5ALB
 
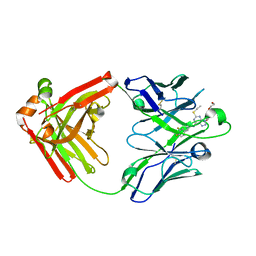 | | Ticagrelor antidote candidate MEDI2452 in complex with ticagrelor | | Descriptor: | MEDI2452 HEAVY CHAIN, MEDI2452 LIGHT CHAIN, Ticagrelor | | Authors: | Buchanan, A, Newton, P, Pehrsson, S, Inghardt, T, Antonsson, T, Svensson, P, Sjogren, T, Oster, L, Janefeldt, A, Sandinge, A, Keyes, F, Austin, M, Spooner, J, Penney, M, Howells, G, Vaughan, T, Nylander, S. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural and Functional Characterisation of a Specific Antidote for Ticagrelor.
Blood, 125, 2015
|
|
5CTQ
 
 | |
5DIE
 
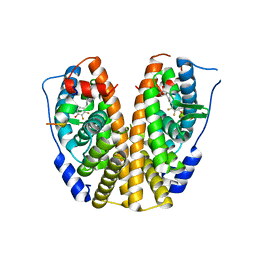 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a trifluoro-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-7a-methyl-5-(2,3,5-trifluoro-4-hydroxyphenyl)octahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-7a-methyl-5-(2,3,5-trifluoro-4-hydroxyphenyl)octahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5D1L
 
 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 (Y165A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, OXALATE ION, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
