9K2D
 
 | | Structure of ClpP from Staphylococcus aureus in complex with ZY39 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6~{S},9~{a}~{S})-6-[(2~{S})-butan-2-yl]-8-[(4-methoxynaphthalen-1-yl)methyl]-4,7-bis(oxidanylidene)-~{N}-[4,4,4-tris(fluoranyl)butyl]-3,6,9,9~{a}-tetrahydro-2~{H}-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Wei, B.Y, Wang, P.Y, Wu, W, Zhang, T, Yang, C.-G. | | Deposit date: | 2024-10-17 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Guided Development of ClpP Agonists with Potent Therapeutic Activities against Staphylococcus aureus Infection.
J.Med.Chem., 68, 2025
|
|
6OOH
 
 | | CTX-M-27 Beta Lactamase with Compound 14 | | Descriptor: | 3-(1H-pyrazol-1-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
4BM8
 
 | | Galectin-3c in complex with Bisamido-thiogalactoside derivate 3 | | Descriptor: | (3-Deoxy-3-(2,3,5,6-tetra-fluoro-4-methoxy-benzamido)-b-D-galactopyranosyl)-(3-deoxy-3-(2,3,5,6-tetra-fluoro-4-methoxy-benzamido)-2-O-sulfo-b-D-galactopyranosyl)-sulfide, GALECTIN-3 | | Authors: | Noresson, A.L, Oberg, C.T, Engstrom, O, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2013-05-07 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Controlling Protein Conformation Through Electronic Fine-Tuning of Arginine-Arene Interactions: Synthetic, Structural, and Biological Studies
To be Published
|
|
5DKQ
 
 | | Crystal Structure of Calcium-loaded S100B bound to SBi4214 | | Descriptor: | 2,2'-[pentane-1,5-diylbis(oxybenzene-4,1-diyl)]di-1,4,5,6-tetrahydropyrimidine, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
9K2B
 
 | | Structure of ClpP from Staphylococcus aureus in complex with ZY18 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (6~{S},9~{a}~{S})-6-[(2~{S})-butan-2-yl]-~{N}-[(2~{S})-2-methylbutyl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-3,6,9,9~{a}-tetrahydro-2~{H}-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wei, B.Y, Wang, P.Y, Wu, W, Zhang, T, Yang, C.-G. | | Deposit date: | 2024-10-17 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Guided Development of ClpP Agonists with Potent Therapeutic Activities against Staphylococcus aureus Infection.
J.Med.Chem., 68, 2025
|
|
5ZU3
 
 | | Effect of mutation (R554K) on FAD modification in Aspergillus oryzae RIB40formate oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Formate oxidase, ... | | Authors: | Mikami, B, Uchida, H, Doubayashi, D. | | Deposit date: | 2018-05-06 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The microenvironment surrounding FAD mediates its conversion to 8-formyl-FAD in Aspergillus oryzae RIB40 formate oxidase.
J.Biochem., 166, 2019
|
|
6VUY
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT358 | | Descriptor: | (7S)-7-phenyl-2-{[3-(piperidin-1-yl)propyl]sulfanyl}-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-amine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
5HTC
 
 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3372 | | Descriptor: | (2R)-2-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}butanedioic acid (non-preferred name), (4S)-2-METHYL-2,4-PENTANEDIOL, ARC-3372 INHIBITOR, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5FVR
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-METHYL-6-(2-(5-(1-METHYLPIPERIDIN-4-YL)PYRIDIN-3-YL) ETHYL)PYRIDIN-2-AMINE | | Descriptor: | 4-methyl-6-(2-(5-(1-methylpiperidin-4-yl)pyridin-3-yl)ethyl)pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2016-02-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibition by Optimization of the 2-Aminopyridine-Based Scaffold with a Pyridine Linker.
J.Med.Chem., 59, 2016
|
|
6SZY
 
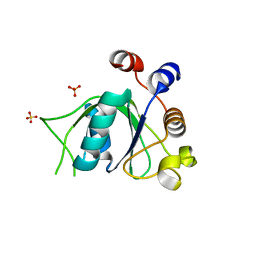 | | Crystal structure of YTHDC1 with fragment 12 (DHU_DC1_150) | | Descriptor: | 4-propan-2-yl-5,6,7,8-tetrahydro-4~{a}~{H}-quinazolin-2-one, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
4GD7
 
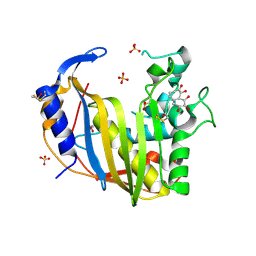 | | Wild-Type Human Thymidylate Synthase with bound Purpurogallin | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,6-tetrahydroxy-5H-benzo[7]annulen-5-one, SULFATE ION, ... | | Authors: | Celeste, L.R, Lebioda, L. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Oxidation of Cysteine 195 of Huyman Thymidylate Synthase by Purpurogallin
To be published, 2012
|
|
6AV2
 
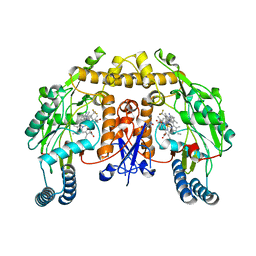 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(3-(3-(dimethylamino)propyl)-5-(trifluoromethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[3-(dimethylamino)propyl]-5-(trifluoromethyl)phenyl}ethyl)-4-methylpyridin-2-amine, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
6NGS
 
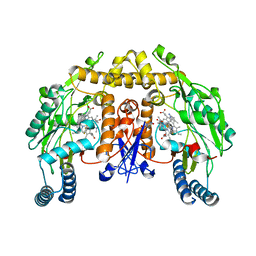 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(5-(3-(dimethylamino)propyl)-2,3-difluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[3-(dimethylamino)propyl]-2,3-difluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
6NH4
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 6-(3-fluoro-5-(2-((2R,4S)-4-fluoropyrrolidin-2-yl)ethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(3-fluoro-5-{2-[(2R,4S)-4-fluoropyrrolidin-2-yl]ethyl}phenyl)ethyl]-4-methylpyridin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
8TXO
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound to the unnatural dZ-PTP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Aiyer, S, Shan, Z, Oh, J, Wang, D, Tan, Y.Z, Lyumkis, D. | | Deposit date: | 2023-08-23 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Overcoming resolution attenuation during tilted cryo-EM data collection.
Nat Commun, 15, 2024
|
|
6SZ3
 
 | | Crystal structure of YTHDC1 with fragment 4 (DHU_DC1_158) | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N}-methyl-5,6,7,8-tetrahydroquinolin-4-amine | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
5WM9
 
 | |
8I69
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Fluoroorotic acid and Citric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CITRIC ACID, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
6AUT
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(3-(3-(Dimethylamino)propyl)-5-fluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[3-(dimethylamino)propyl]-5-fluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
8R44
 
 | | PAS-GAF bidomain of Glycine max phytochrome A | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, PHYCOCYANOBILIN, ... | | Authors: | Guan, K, Nagano, S, Hughes, J. | | Deposit date: | 2023-11-13 | | Release date: | 2025-01-08 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Pr and Pfr structures of plant phytochrome A.
Nat Commun, 16, 2025
|
|
8I65
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with isoorotic acid (2,4-Dihydroxypyrimidine-5-carboxylic Acid), Form I | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I66
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with isoorotic acid (2,4-Dihydroxypyrimidine-5-carboxylic Acid) and citric acid, Form I | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, CITRIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8U2E
 
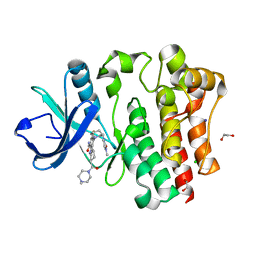 | | Bruton's tyrosine kinase in complex with N-[(2R)-1-[(3R)-3-(methylcarbamoyl)-1H,2H,3H,4H,9H-pyrido[3,4-b]indol-2-yl]-3-(3-methylphenyl)-1-oxopropan-2-yl]-1H-indazole-5-carboxamide | | Descriptor: | (2S)-6-fluoro-5-[(3S)-3-(3-methyl-2-oxoimidazolidin-1-yl)piperidin-1-yl]-2-(4-methylpiperazine-1-carbonyl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Gajewski, S, Clifton, M.C. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Preclinical Pharmacology of NX-2127, an Orally Bioavailable Degrader of Bruton's Tyrosine Kinase with Immunomodulatory Activity for the Treatment of Patients with B Cell Malignancies.
J.Med.Chem., 67, 2024
|
|
3ZM6
 
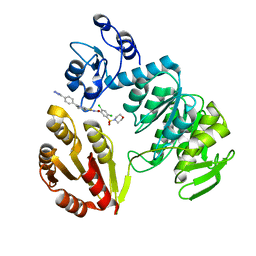 | | CRYSTAL STRUCTURE OF MURF LIGASE IN COMPLEX WITH CYANOTHIOPHENE INHIBITOR | | Descriptor: | N-(6-(4-(2h-tetrazol-5-yl)benzyl)-3-cyano-4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl)-2,4-dichloro-5-(morpholinosulfonyl)benzamide, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | Authors: | Hrast, M, Turk, S, Sosic, I, Knez, D, Randall, C.P, Barreteau, H, Contreras-Martel, C, Dessen, A, ONeill, A.J, Mengin-Lecreulx, D, Blanot, D, Gobec, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Activity Relationships of New Cyanothiophene Inhibitors of the Essential Peptidoglycan Biosynthesis Enzyme Murf.
Eur.J.Med.Chem., 66C, 2013
|
|
8U2D
 
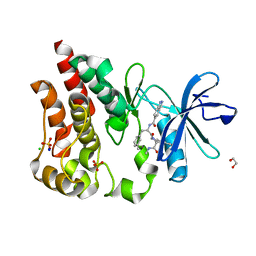 | | Bruton's tyrosine kinase in complex with N-[(2R)-1-[(3R)-3-(methylcarbamoyl)-1H,2H,3H,4H,9H-pyrido[3,4-b]indol-2-yl]-3-(3-methylphenyl)-1-oxopropan-2-yl]-1H-indazole-5-carboxamide | | Descriptor: | (3R)-2-[N-(1H-indazole-5-carbonyl)-3-methyl-D-phenylalanyl]-N-methyl-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Gajewski, S, Clifton, M.C. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Preclinical Pharmacology of NX-2127, an Orally Bioavailable Degrader of Bruton's Tyrosine Kinase with Immunomodulatory Activity for the Treatment of Patients with B Cell Malignancies.
J.Med.Chem., 67, 2024
|
|
