8HYI
 
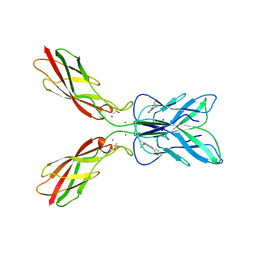 | | Crystal structure of human P-cadherin MEC12 (X dimer) in complex with 2-(2-methyl-5-phenyl-1H-indole-3-yl)ethan-1-amine | | Descriptor: | 2-(2-methyl-5-phenyl-1H-indole-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3, ... | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2023-01-06 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Modulation of a conformational ensemble by a small molecule that inhibits key protein-protein interactions involved in cell adhesion.
Protein Sci., 32, 2023
|
|
4UPJ
 
 | |
3V3V
 
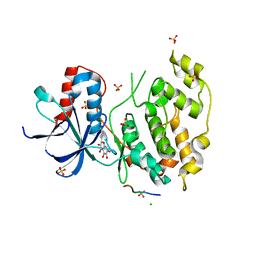 | | Structural and functional analysis of quercetagetin, a natural JNK1 inhibitor | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, C-Jun-amino-terminal kinase-interacting protein 1, CHLORIDE ION, ... | | Authors: | Baek, S, Kang, N.J, Popowicz, G.M, Arciniega, M, Jung, S.K, Byun, S, Song, N.R, Heo, Y.S, Kim, B.Y, Lee, H.J, Holak, T.A, Augustin, M, Bode, A.M, Huber, R, Dong, Z, Lee, K.W. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of the Natural JNK1 Inhibitor Quercetagetin.
J.Mol.Biol., 425, 2013
|
|
1SME
 
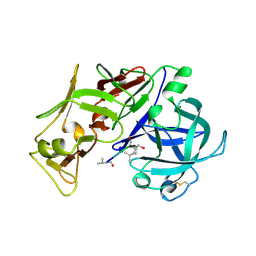 | | PLASMEPSIN II, A HEMOGLOBIN-DEGRADING ENZYME FROM PLASMODIUM FALCIPARUM, IN COMPLEX WITH PEPSTATIN A | | Descriptor: | PLASMEPSIN II, Pepstatin | | Authors: | Silva, A.M, Lee, A.Y, Gulnik, S.V, Goldberg, D.E, Erickson, J.W. | | Deposit date: | 1996-06-11 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1SL4
 
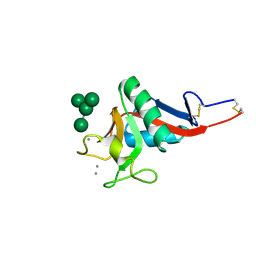 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with Man4 | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, mDC-SIGN1B type I isoform | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
6K59
 
 | |
4UNG
 
 | | Human insulin B26Asn mutant crystal structure | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN, SULFATE ION | | Authors: | Zakova, L, Klevtikova, E, Lepsik, M, Collinsova, M, Watson, C.J, Turkenburg, J.P, Jiracek, J, Brzozowski, A.M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Human Insulin Analogues Modified at the B26 Site Reveal a Hormone Conformation that is Undetected in the Receptor Complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1SPR
 
 | |
1SN7
 
 | | KUMAMOLISIN-AS, APOENZYME | | Descriptor: | CALCIUM ION, kumamolisin-As | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Oda, K, Nishino, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-as, a serine-carboxyl peptidase with collagenase activity.
J.Biol.Chem., 279, 2004
|
|
6KAH
 
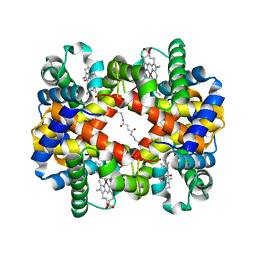 | | Crosslinked alpha(Ni)-beta(Fe-CO) human hemoglobin A in the T quaternary structure at 95 K: Dark | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-23 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KAQ
 
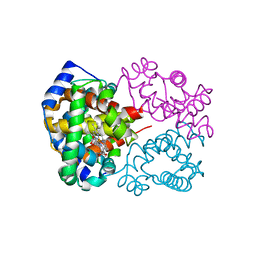 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 140 K: Dark | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KAU
 
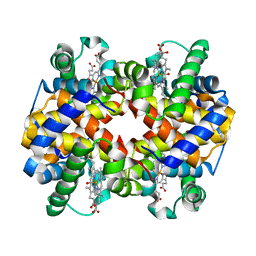 | | Carbonmonoxy human hemoglobin A in the R2 quaternary structure at 140 K: Dark | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1SOS
 
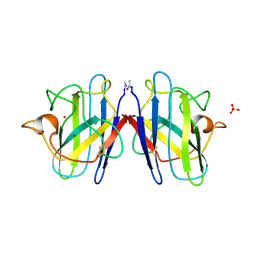 | | ATOMIC STRUCTURES OF WILD-TYPE AND THERMOSTABLE MUTANT RECOMBINANT HUMAN CU, ZN SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE, ... | | Authors: | Parge, H.E, Hallewell, R.A, Tainer, J.A. | | Deposit date: | 1992-02-11 | | Release date: | 1993-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structures of wild-type and thermostable mutant recombinant human Cu,Zn superoxide dismutase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
3UVP
 
 | | Human p38 MAP Kinase in Complex with a Benzamide Substituted Benzosuberone | | Descriptor: | Mitogen-activated protein kinase 14, N-{2-fluoro-5-[(5-oxo-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)amino]phenyl}benzamide, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Richters, A, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Hinge Glycine Flip and the Activation Loop: Novel Approach to Potent p38 alpha Inhibitors.
J.Med.Chem., 55, 2012
|
|
1SQO
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 8-(PYRIMIDIN-2-YLAMINO)NAPHTHALENE-2-CARBOXIMIDAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhao, X, Mantei, R, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-19 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
3UNS
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UNX
 
 | | Bond length analysis of asp, glu and his residues in subtilisin Carlsberg at 1.26A resolution | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Fisher, S.J, Helliwell, J.R, Blakeley, M.P, Cianci, M, McSweeny, S. | | Deposit date: | 2011-11-16 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Protonation-state determination in proteins using high-resolution X-ray crystallography: effects of resolution and completeness.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1SQZ
 
 | | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between Group II Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution | | Descriptor: | Phospholipase A2, SULFATE ION, synthetic peptide | | Authors: | Singh, N, Prem Kumar, R, Somvanshi, R.K, Bilgrami, S, Ethayathulla, A.S, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between GroupII Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution
To be Published
|
|
3UOP
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-17 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
8IBX
 
 | | Structure of R2 with 3'UTR and DNA in unwinding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8SDK
 
 | | The MicroED structure of proteinase K crystallized by suspended drop crystallization | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Gillman, C, Nicolas, W.J, Martynowycz, M.W, Gonen, T. | | Deposit date: | 2023-04-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | Design and implementation of suspended drop crystallization.
Iucrj, 10, 2023
|
|
4UHL
 
 | | HUMAN STEROL 14-ALPHA DEMETHYLASE (CYP51) IN COMPLEX WITH VFV IN P1 SPACE GROUP | | Descriptor: | N-[(1R)-1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, I Lepesheva, G. | | Deposit date: | 2015-03-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human Sterol 14Alpha-Demethylase (Cyp51) as a Target for Anticancer Chemotherapy: Towards Structure-Aided Drug Design.
J.Lipid Res., 57, 2016
|
|
3V1R
 
 | | Crystal structures of the reverse transcriptase-associated ribonuclease H domain of XMRV with inhibitor beta-thujaplicinol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, MANGANESE (II) ION, ... | | Authors: | Zhou, D, Wlodawer, A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the reverse transcriptase-associated ribonuclease H domain of xenotropic murine leukemia-virus related virus.
J.Struct.Biol., 177, 2012
|
|
7BZW
 
 | | Structure of Hsp21 | | Descriptor: | Heat shock protein 21, chloroplastic | | Authors: | Lau, W.C.Y. | | Deposit date: | 2020-04-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of substrate recognition and thermal protection by a small heat shock protein.
Nat Commun, 12, 2021
|
|
7BFL
 
 | | X-ray structure of SS-RNase-2 des116-120 | | Descriptor: | Angiogenin-1 | | Authors: | Sica, F, Russo Krauss, I, Troisi, R. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | The structural features of an ancient ribonuclease from Salmo salar reveal an intriguing case of auto-inhibition.
Int.J.Biol.Macromol., 182, 2021
|
|
