1IH3
 
 | |
1IH4
 
 | |
1IH5
 
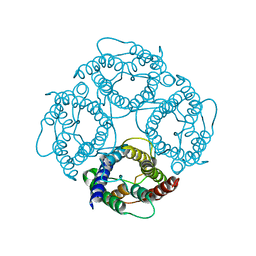 | | CRYSTAL STRUCTURE OF AQUAPORIN-1 | | Descriptor: | AQUAPORIN-1 | | Authors: | Ren, G, Reddy, V.S, Cheng, A, Melnyk, P, Mitra, A.K. | | Deposit date: | 2001-04-18 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.7 Å) | | Cite: | Visualization of a water-selective pore by electron crystallography in vitreous ice.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IH6
 
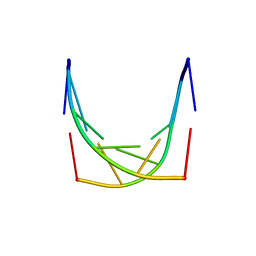 | |
1IH7
 
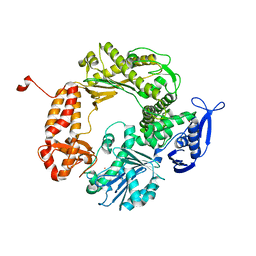 | | High-Resolution Structure of Apo RB69 DNA Polymerase | | Descriptor: | DNA POLYMERASE, GUANOSINE, POTASSIUM ION | | Authors: | Franklin, M.C, Wang, J, Steitz, T.A. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the Replicating Complex of a Pol alpha Family DNA Polymerase
Cell(Cambridge,Mass.), 105, 2001
|
|
1IH8
 
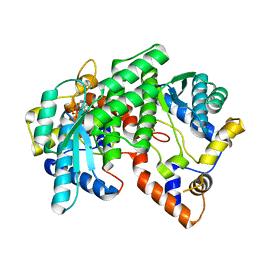 | | NH3-dependent NAD+ Synthetase from Bacillus subtilis Complexed with AMP-CPP and Mg2+ ions. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) synthetase | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, DeLucas, L. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IH9
 
 | | NMR Structure of Zervamicin IIB (peptaibol antibiotic) Bound to DPC Micelles | | Descriptor: | ZERVAMICIN IIB | | Authors: | Shenkarev, Z.O, Balasheva, T.A, Efremov, R.G, Yakimenko, Z.A, Ovchinnikova, T.V, Raap, J, Arseniev, A.S. | | Deposit date: | 2001-04-19 | | Release date: | 2002-02-13 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of Zervamicin Iib Bound to Dpc Micelles: Implications for Voltage-Gating.
Biophys.J., 82, 2002
|
|
1IHA
 
 | | Structure of the Hybrid RNA/DNA R-GCUUCGGC-D[BR]U in Presence of RH(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(BRU))-3', CHLORIDE ION, RHODIUM HEXAMINE ION | | Authors: | Cruse, W.B, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IHB
 
 | | CRYSTAL STRUCTURE OF P18-INK4C(INK6) | | Descriptor: | CYCLIN-DEPENDENT KINASE 6 INHIBITOR | | Authors: | Ravichandran, V, Swaminathan, K, Marmorstein, R. | | Deposit date: | 1997-10-25 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the CDK4/6 inhibitory protein p18INK4c provides insights into ankyrin-like repeat structure/function and tumor-derived p16INK4 mutations.
Nat.Struct.Biol., 5, 1998
|
|
1IHC
 
 | | X-ray Structure of Gephyrin N-terminal Domain | | Descriptor: | Gephyrin | | Authors: | Sola, M, Kneussel, M, Heck, I.S, Betz, H, Weissenhorn, W. | | Deposit date: | 2001-04-21 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of the trimeric N-terminal domain of gephyrin.
J.Biol.Chem., 276, 2001
|
|
1IHD
 
 | |
1IHF
 
 | | INTEGRATION HOST FACTOR/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (35-MER), DNA (5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*C P*AP*CP*C)-3'), ... | | Authors: | Rice, P.A, Yang, S.-W, Mizuuchi, K, Nash, H.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn.
Cell(Cambridge,Mass.), 87, 1996
|
|
1IHG
 
 | | Bovine Cyclophilin 40, monoclinic form | | Descriptor: | Cyclophilin 40, GLYCEROL | | Authors: | Taylor, P, Dornan, J, Carrello, A, Minchin, R.F, Ratajczak, T, Walkinshaw, M.D. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two structures of cyclophilin 40: folding and fidelity in the TPR domains.
Structure, 9, 2001
|
|
1IHH
 
 | | 2.4 ANGSTROM CRYSTAL STRUCTURE OF AN OXALIPLATIN 1,2-D(GPG) INTRASTRAND CROSS-LINK IN A DNA DODECAMER DUPLEX | | Descriptor: | 1R,2R-DIAMINOCYCLOHEXANE, 5'-D(*CP*CP*TP*CP*TP*GP*GP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3', ... | | Authors: | Spingler, B, Whittington, D.A, Lippard, S.J. | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 A crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand cross-link in a DNA dodecamer duplex.
Inorg.Chem., 40, 2001
|
|
1IHI
 
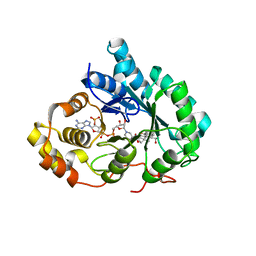 | | Crystal Structure of Human Type III 3-alpha-Hydroxysteroid Dehydrogenase/Bile Acid Binding Protein (AKR1C2) Complexed with NADP+ and Ursodeoxycholate | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, ISO-URSODEOXYCHOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jin, Y, Stayrook, S.E, Albert, R.H, Palackal, N.T, Penning, T.M, Lewis, M. | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human type III 3alpha-hydroxysteroid dehydrogenase/bile acid binding protein complexed with NADP(+) and ursodeoxycholate.
Biochemistry, 40, 2001
|
|
1IHJ
 
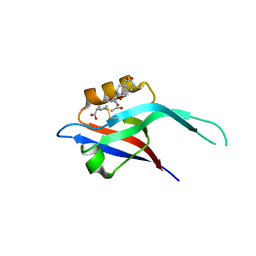 | |
1IHK
 
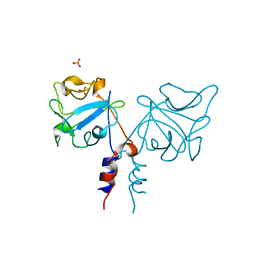 | | CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 9 (FGF9) | | Descriptor: | GLIA-ACTIVATING FACTOR, PHOSPHATE ION | | Authors: | Plotnikov, A.N, Eliseenkova, A.V, Ibrahimi, O.A, Lemmon, M.A, Mohammadi, M. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of fibroblast growth factor 9 reveals regions implicated in dimerization and autoinhibition.
J.Biol.Chem., 276, 2001
|
|
1IHM
 
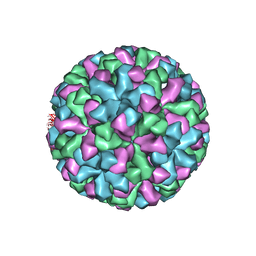 | | CRYSTAL STRUCTURE ANALYSIS OF NORWALK VIRUS CAPSID | | Descriptor: | capsid protein | | Authors: | Prasad, B.V, Hardy, M.E, Dokland, T, Bella, J, Rossmann, M.G, Estes, M.K. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray crystallographic structure of the Norwalk virus capsid
Science, 286, 1999
|
|
1IHN
 
 | |
1IHO
 
 | | CRYSTAL APO-STRUCTURE OF PANTOTHENATE SYNTHETASE FROM E. COLI | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PANTOATE--BETA-ALANINE LIGASE | | Authors: | von Delft, F, Lewendon, A, Dhanaraj, V, Blundell, T.L, Abell, C, Smith, A. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of E. coli pantothenate synthetase confirms it as a member of the cytidylyltransferase superfamily.
Structure, 9, 2001
|
|
1IHP
 
 | | STRUCTURE OF PHOSPHOMONOESTERASE | | Descriptor: | PHYTASE, SULFATE ION | | Authors: | Kostrewa, D. | | Deposit date: | 1997-02-04 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of phytase from Aspergillus ficuum at 2.5 A resolution.
Nat.Struct.Biol., 4, 1997
|
|
1IHQ
 
 | | GLYTM1BZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF A RAT SHORT ALPHA TROPOMYOSIN WITH THE N-TERMINUS ENCODED BY EXON 1B | | Descriptor: | CHIMERIC PEPTIDE GlyTM1bZip: TROPOMYOSIN ALPHA CHAIN, BRAIN-3 and GENERAL CONTROL PROTEIN GCN4 | | Authors: | Greenfield, N.J, Yuang, Y.J, Palm, T, Swapna, G.V, Monleon, D, Montelione, G.T, Hitchcock-Degregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and folding dynamics of the N terminus of a rat non-muscle alpha-tropomyosin in an engineered chimeric protein.
J.Mol.Biol., 312, 2001
|
|
1IHR
 
 | | Crystal structure of the dimeric C-terminal domain of TonB | | Descriptor: | BROMIDE ION, TonB protein | | Authors: | Chang, C, Mooser, A, Pluckthun, A, Wlodawer, A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold.
J.Biol.Chem., 276, 2001
|
|
1IHS
 
 | |
1IHT
 
 | |
