8HNP
 
 | | Archaeal transcription factor Mutant | | Descriptor: | Archaeal transcription regulator | | Authors: | Bae, D.W, Cha, S.S. | | Deposit date: | 2022-12-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | An archaeal transcription factor EnfR with a novel 'eighth note' fold controls hydrogen production of a hyperthermophilic archaeon Thermococcus onnurineus NA1.
Nucleic Acids Res., 51, 2023
|
|
8HNO
 
 | | Archaeal transcription factor Wild type | | Descriptor: | Archaeal transcription regulator | | Authors: | Bae, D.W, Cha, S.S. | | Deposit date: | 2022-12-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | An archaeal transcription factor EnfR with a novel 'eighth note' fold controls hydrogen production of a hyperthermophilic archaeon Thermococcus onnurineus NA1.
Nucleic Acids Res., 51, 2023
|
|
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H2G
 
 | | Cryo-EM structure of niacin bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2
Nat Commun, 14, 2023
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.51 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3QAM
 
 | | Crystal Structure of Glu208Ala mutant of catalytic subunit of cAMP-dependent protein kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | Authors: | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | Deposit date: | 2011-01-11 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
8H36
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7VH4
 
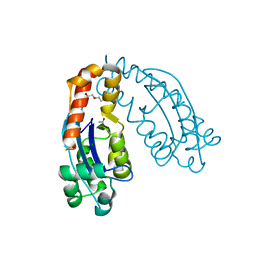 | | Crystal structure of oligoribonuclease of Escherichia coli | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Badhwar, P, Taneja, B. | | Deposit date: | 2021-09-20 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of a mycobacterial oligoribonuclease reveals a unique C-terminal tail that stabilizes the homodimer.
J.Biol.Chem., 298, 2022
|
|
8H37
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.52 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3SUO
 
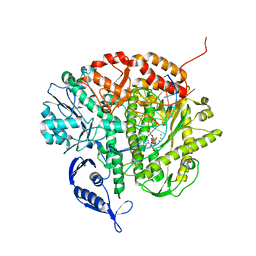 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dTTP Opposite 2AP (GC rich sequence) | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
7VF3
 
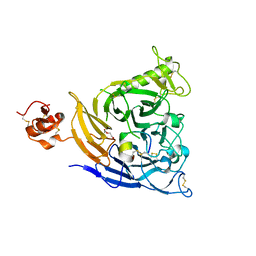 | | Plexin B1 extracellular fragment in complex with lasso-grafted PB1m7 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Plexin-B1, ... | | Authors: | Sugano, N.N, Hirata, K, Yamashita, K, Yamamoto, M, Arimori, T, Takagi, J. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | De novo Fc-based receptor dimerizers differentially modulate PlexinB1 function.
Structure, 30, 2022
|
|
7VS3
 
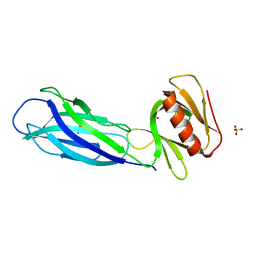 | | The crystal structure of rat calcium-dependent activator protein for secretion (CAPS) C2PH | | Descriptor: | Calcium-dependent secretion activator 1, SULFATE ION | | Authors: | Zhou, H, Wei, Z.Q, Zhang, L, Ren, Y.J, Ma, C. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | The C 2 and PH domains of CAPS constitute an effective PI(4,5)P2-binding unit essential for Ca 2+ -regulated exocytosis.
Structure, 31, 2023
|
|
8H92
 
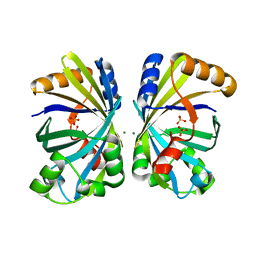 | |
8HLP
 
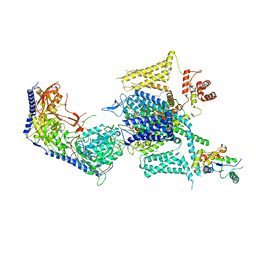 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 (apo) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8GYQ
 
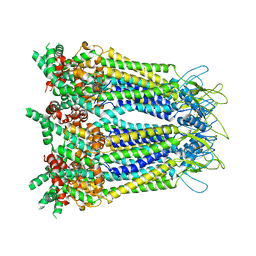 | |
8HMB
 
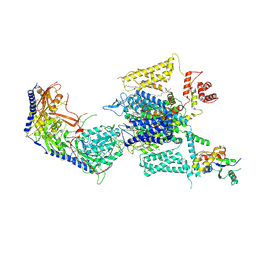 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with benidipine (BEN) | | Descriptor: | (3R)-1-benzylpiperidin-3-yl methyl (2R,3R,4R,5R,6S)-2,6-dimethyl-4-(3-nitrophenyl)piperidine-3,5-dicarboxylate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HMA
 
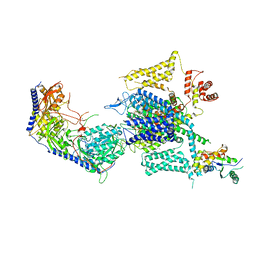 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with tetrandrine (TET) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6,6',7,12-tetramethoxy-2,2'-dimethyl-1beta-3,4-didehydroberbaman, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
7W5O
 
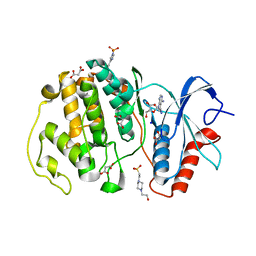 | | Crystal structure of ERK2 with an allosteric inhibitor | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 13-[4-({Imidazo[1,2-a]pyridin-2-yl}methoxy)phenyl]-4,8-dioxa-12,14,16,18-tetraazatetracyclo[9.7.0.0^{3,9}.0^{12,17}]octadeca-1(11),2,9,15,17-pentaen-15-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of a novel target site for ATP-independent ERK2 inhibitors.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
3SUN
 
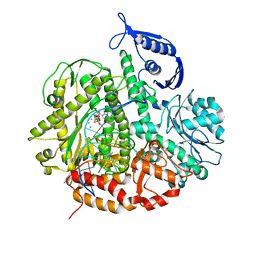 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dTTP Opposite 2AP (AT rich sequence) | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
3POO
 
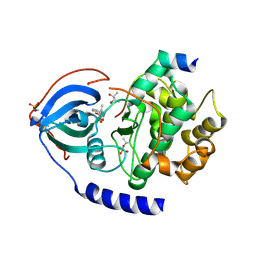 | | human cAMP-dependent protein kinase in complex with an inhibitor | | Descriptor: | (2S,3S)-butane-2,3-diol, N'-[(E)-(2,4-dihydroxy-6-methylphenyl)methylidene]-2-(3-methoxyphenyl)acetohydrazide, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-23 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | fragment based drug design on PKA
To be Published
|
|
8IG0
 
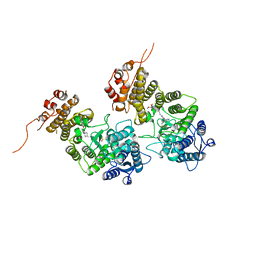 | | Crystal structure of menin in complex with DS-1594b | | Descriptor: | (1R,2S,4R)-4-[[4-(5,6-dimethoxypyridazin-3-yl)phenyl]methylamino]-2-[methyl-[6-[2,2,2-tris(fluoranyl)ethyl]thieno[2,3-d]pyrimidin-4-yl]amino]cyclopentan-1-ol, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Suzuki, M, Yoneyama, T, Imai, E. | | Deposit date: | 2023-02-20 | | Release date: | 2023-03-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel Menin-MLL1 inhibitor, DS-1594a, prevents the progression of acute leukemia with rearranged MLL1 or mutated NPM1.
Cancer Cell Int, 23, 2023
|
|
3PVB
 
 | | Crystal structure of (73-244)RIa:C holoenzyme of cAMP-dependent Protein kinase | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Boettcher, A.J, Wu, J, Kim, C, Yang, J, Bruystens, J, Cheung, N, Pennypacker, J.K, Blumenthal, D.A, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2010-12-06 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of (73-244)RIa:C holoenzyme of cAMP-dependent Protein kinase
Structure, 19, 2011
|
|
8IJN
 
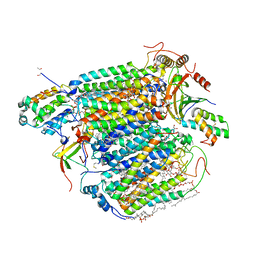 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-Bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A, Muramoto, K. | | Deposit date: | 2023-02-27 | | Release date: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8HUJ
 
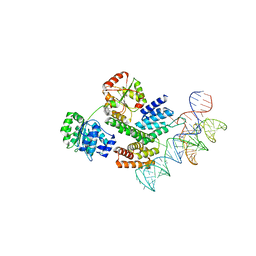 | | Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A | | Descriptor: | Eukaryotic initiation factor 4A-I, Eukaryotic translation initiation factor 4 gamma 1, IRES RNA (J-K-St), ... | | Authors: | Suzuki, H, Fujiyoshi, Y, Imai, S, Shimada, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Dynamically regulated two-site interaction of viral RNA to capture host translation initiation factor.
Nat Commun, 14, 2023
|
|
8I92
 
 | | ACE2-B0AT1 complex bound with glutamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
