4I5I
 
 | | Crystal structure of the SIRT1 catalytic domain bound to NAD and an EX527 analog | | Descriptor: | (6S)-2-chloro-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhao, X, Allison, D, Condon, B, Zhang, F, Gheyi, T, Zhang, A, Ashok, S, Russell, M, Macewan, I, Qian, Y, Jamison, J.A, Luz, J.G. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 angstrom crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition.
J.Med.Chem., 56, 2013
|
|
2I39
 
 | | Crystal structure of Vaccinia virus N1L protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Protein N1 | | Authors: | Aoyagi, M, Aleshin, A.E, Stec, B, Liddington, R.C. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vaccinia virus N1L protein resembles a B cell lymphoma-2 (Bcl-2) family protein.
Protein Sci., 16, 2007
|
|
1COW
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AUROVERTIN B, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | van Raaij, M.J, Abrahams, J.P, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-08 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
4LVF
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | (1S,2S)-2-phenyl-N-(pyridin-4-yl)cyclopropanecarboxamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1D3T
 
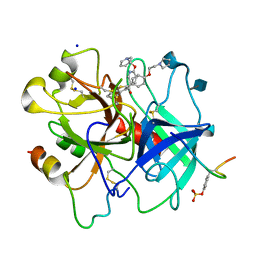 | |
1D3P
 
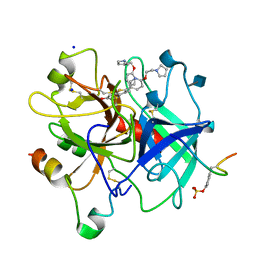 | | CRYSTAL STRUCTURE OF HUMAN APLHA-THROMBIN IN COMPLEX WITH BENZO[B]THIOPHENE INHIBITOR 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-(2-PYRROLIDIN-1-YL-ETHOXY)-BENZYL]-2-4-(2-PYRROLIDIN-1-YL-ETHOXY)-PHENYL] -BENZO[B]THIOPHEN-6-OL, ALPHA-THROMBIN, ... | | Authors: | Chirgadze, N.Y. | | Deposit date: | 1999-09-30 | | Release date: | 2000-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of human alpha-thrombin complexed with active site-directed diamino benzo[b]thiophene derivatives: a binding mode for a structurally novel class of inhibitors.
Protein Sci., 9, 2000
|
|
6CHP
 
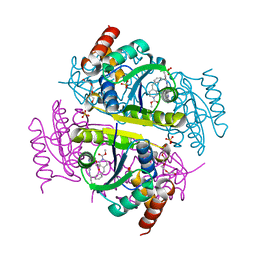 | |
4LVA
 
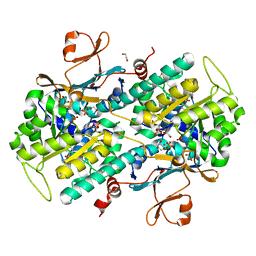 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[4-(pyrrolidin-1-yl)piperidin-1-yl]sulfonyl}benzyl)-2H-pyrido[4,3-e][1,2,4]thiadiazin-3-amine 1,1-dioxide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HZ5
 
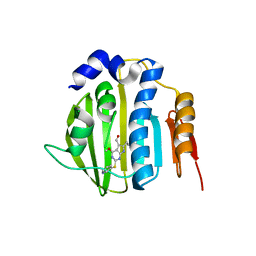 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity | | Descriptor: | 6-ethyl-4-methoxy-2-(pyridin-3-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbaldehyde, DNA topoisomerase IV, B subunit | | Authors: | Bensen, D.C, Creighton, C.J, Tari, L.W. | | Deposit date: | 2012-11-14 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3KPR
 
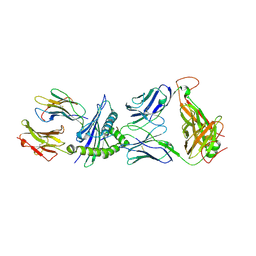 | | Crystal Structure of the LC13 TCR in complex with HLA B*4405 bound to EEYLKAWTF a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KQ4
 
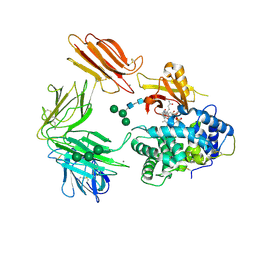 | | Structure of Intrinsic Factor-Cobalamin bound to its receptor Cubilin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Andersen, C.B.F, Madsen, M, Moestrup, S.K, Andersen, G.R. | | Deposit date: | 2009-11-17 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes.
Nature, 464, 2010
|
|
3KPM
 
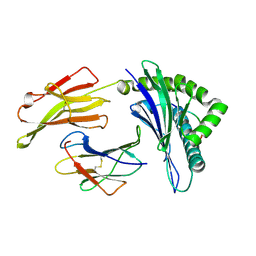 | | Crystal Structure of HLA B*4402 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
1AN2
 
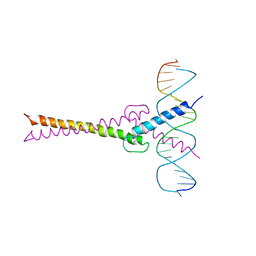 | | RECOGNITION BY MAX OF ITS COGNATE DNA THROUGH A DIMERIC B/HLH/Z DOMAIN | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*GP*GP*TP*CP*AP*CP*GP*TP*GP*AP*CP*C P*TP*AP*CP*AP*C)- 3'), PROTEIN (TRANSCRIPTION FACTOR MAX (TF MAX)) | | Authors: | Ferre-D'Amare, A.R, Prendergast, G.C, Ziff, E.B, Burley, S.K. | | Deposit date: | 1996-09-06 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain.
Nature, 363, 1993
|
|
4J6Q
 
 | |
1E6N
 
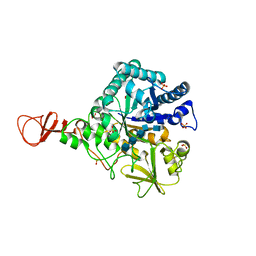 | | Chitinase B from Serratia marcescens inactive mutant E144Q in complex with N-acetylglucosamine-pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE B, GLYCEROL, ... | | Authors: | Komander, D, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2000-08-21 | | Release date: | 2001-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of a Family 18 Exo-Chitinase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GPY
 
 | |
3KPP
 
 | | Crystal Structure of HLA B*4405 in complex with EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, EEYLQAFTY, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
1E97
 
 | |
3KPQ
 
 | | Crystal Structure of HLA B*4405 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KPL
 
 | | Crystal Structure of HLA B*4402 in complex with EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KPS
 
 | | Crystal Structure of the LC13 TCR in complex with HLA B*4405 bound to EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
1D3Q
 
 | | CRYSTAL STRUCTURE OF HUMAN ALPHA THROMBIN IN COMPLEX WITH BENZO[B]THIOPHENE INHIBITOR 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-(2-PYRROLIDIN-1-YL-ETHOXY)-BENZYL]-2-4-(2-PYRROLIDIN-1-YL-ETHOXY)-PHENYL] -BENZO[B]THIOPHENE, ALPHA-THROMBIN, ... | | Authors: | Chirgadze, N.Y. | | Deposit date: | 1999-09-30 | | Release date: | 2000-10-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structures of human alpha-thrombin complexed with active site-directed diamino benzo[b]thiophene derivatives: a binding mode for a structurally novel class of inhibitors.
Protein Sci., 9, 2000
|
|
4K7F
 
 | | Newly identified epitope V60 from HBV core protein complexed with HLA-A*0201 | | Descriptor: | Beta-2-microglobulin, Core protein, HLA class I histocompatibility antigen, ... | | Authors: | Meng, S.D, Zhang, Y, Wu, Y, Qi, J.X. | | Deposit date: | 2013-04-17 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The L60V variation in hepatitis B virus core protein elicits new epitope-specific cytotoxic T lymphocytes and enhances viral replication.
J.Virol., 87, 2013
|
|
1E6R
 
 | | Chitinase B from Serratia marcescens wildtype in complex with inhibitor allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE B, ... | | Authors: | Komander, D, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2000-08-22 | | Release date: | 2001-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of a Family 18 Exo-Chitinase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4G96
 
 | | Crystal structure of calcium2+-free wild-type CD23 lectin domain | | Descriptor: | 1,2-ETHANEDIOL, Low affinity immunoglobulin epsilon Fc receptor, SULFATE ION | | Authors: | Yuan, D, Sutton, B.J, Dhaliwal, B. | | Deposit date: | 2012-07-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Ca2+-dependent Structural Changes in the B-cell Receptor CD23 Increase Its Affinity for Human Immunoglobulin E.
J.Biol.Chem., 288, 2013
|
|
