7O4O
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylhomocysteine | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7OCR
 
 | | NADPH and fructose-6-phosphate bound to the dehydrogenase domain of the bifunctional mannitol-1-phosphate dehydrogenase/phosphatase MtlD from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Tam, H.K, Mueller, V, Pos, K.M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unidirectional mannitol synthesis of Acinetobacter baumannii MtlD is facilitated by the helix-loop-helix-mediated dimer formation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7OCN
 
 | | Crystal structure of the bifunctional mannitol-1-phosphate dehydrogenase/phosphatase MtlD from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, ... | | Authors: | Tam, H.K, Mueller, V, Pos, K.M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unidirectional mannitol synthesis of Acinetobacter baumannii MtlD is facilitated by the helix-loop-helix-mediated dimer formation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7O4N
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylmethionine | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O6S
 
 | | Crystal structure of a shortened IpgC variant in complex with N-(2H-1,3-benzodioxol-5-ylmethyl)cyclopentanamine | | Descriptor: | CHLORIDE ION, Chaperone protein IpgC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gardonyi, M, Heine, A, Klebe, G. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a shortened IpgC variant in complex with N-(2H-1,3-benzodioxol-5-ylmethyl)cyclopentanamine
To be published
|
|
7O4M
 
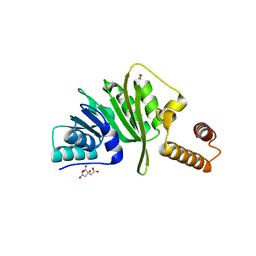 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase | | Descriptor: | CITRIC ACID, GLYCEROL, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7OCP
 
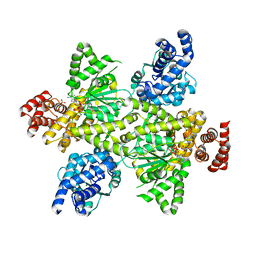 | | NADPH bound to the dehydrogenase domain of the bifunctional mannitol-1-phosphate dehydrogenase/phosphatase MtlD from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Tam, H.K, Mueller, V, Pos, K.M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Unidirectional mannitol synthesis of Acinetobacter baumannii MtlD is facilitated by the helix-loop-helix-mediated dimer formation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7OAC
 
 | | conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta1/A, crystal form I | | Descriptor: | General control transcription factor GCN4,conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta1/A,General control transcription factor GCN4 | | Authors: | Adlakha, J, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta1/A, crystal form I
To Be Published
|
|
7OAF
 
 | | conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta1/A, crystal form III | | Descriptor: | General control transcription factor GCN4,conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta1/A,General control transcription factor GCN4, PENTAETHYLENE GLYCOL | | Authors: | Adlakha, J, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta1/A, crystal form III
To Be Published
|
|
8CMI
 
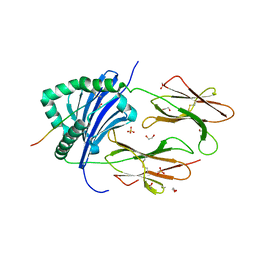 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 Omicron (BA.1) Spike peptide S761-775 | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
8CMH
 
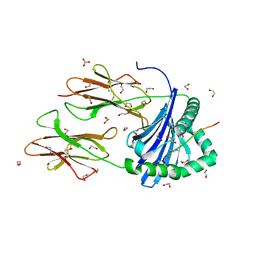 | | Human Leukocyte Antigen class II allotype DR1 presenting SARS-CoV-2 Omicron (BA.1) Spike peptide S486-505 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, HLA class II histocompatibility antigen, ... | | Authors: | MacLachlan, B.J, Mason, G.H, Sourfield, D.O, Godkin, A.J, Rizkallah, P.J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural definition of HLA class II-presented SARS-CoV-2 epitopes reveals a mechanism to escape pre-existing CD4 + T cell immunity.
Cell Rep, 42, 2023
|
|
7OAH
 
 | | conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta2/A | | Descriptor: | General control transcription factor GCN4,conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta2/A,General control transcription factor GCN4, TETRAETHYLENE GLYCOL | | Authors: | Adlakha, J, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta2/A
To Be Published
|
|
7OAD
 
 | | conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta1/A, crystal form II | | Descriptor: | General control transcription factor GCN4,conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta1/A,General control transcription factor GCN4 | | Authors: | Adlakha, J, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | conserved hypothetical protein residues 311-335 from Candidatus Magnetomorum sp. HK-1 fused to GCN4 adaptors, mutant beta1/A, crystal form II
To Be Published
|
|
8CDW
 
 | |
6Q01
 
 | | TDP2 UBA Domain Bound to Ubiquitin at 0.85 Angstroms Resolution, Crystal Form 2 | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Krahn, J.M, Williams, R.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.851 Å) | | Cite: | Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.
Nucleic Acids Res., 48, 2020
|
|
8CHL
 
 | | Human FKBP12 in complex with (1S,5S,6R)-9-((3,5-dichlorophenyl)sulfonyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,9-diazabicyclo[4.2.1]nonan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, CADMIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
7YVW
 
 | | NMR determination of the 2:1 binding motif structure involving cytosine flipping out for the recognition of the CGG/CGG triad DNA | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3') | | Authors: | Furuita, K, Yamada, T, Sakurabayashi, S, Nomura, M, Kojima, C, Nakatani, K. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.
Nucleic Acids Res., 50, 2022
|
|
6QCJ
 
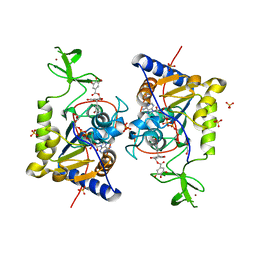 | | Human Sirt6 in complex with ADP-ribose and the inhibitor catechin gallate | | Descriptor: | (2R,3S)-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, 1,2-ETHANEDIOL, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2018-12-28 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives.
Sci Rep, 9, 2019
|
|
6WBK
 
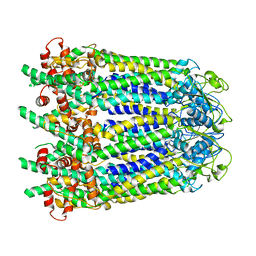 | |
7M67
 
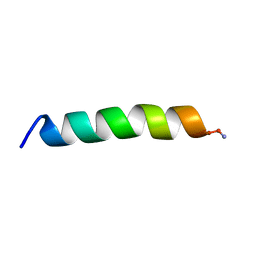 | |
8CCX
 
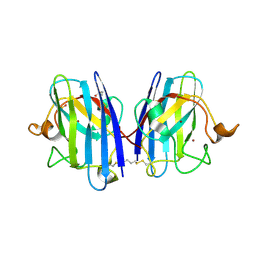 | | Human SOD1 in complex with S-XL6 cross-linker | | Descriptor: | COPPER (II) ION, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Hossain, A, Agar, J.N, Hasnain, S.S. | | Deposit date: | 2023-01-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Evaluating protein cross-linking as a therapeutic strategy to stabilize SOD1 variants in a mouse model of familial ALS.
Plos Biol., 22, 2024
|
|
6Q94
 
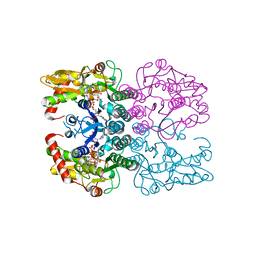 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase (S156D) in complex with GDP-Man | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
7P58
 
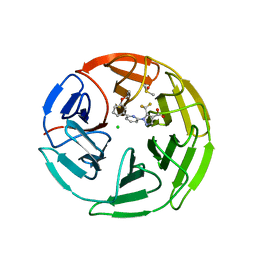 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | Descriptor: | 1-[6-(3-propan-2-yloxyphenyl)pyridin-2-yl]-5-(trifluoromethyl)pyrazole-4-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Davies, T.G. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P5E
 
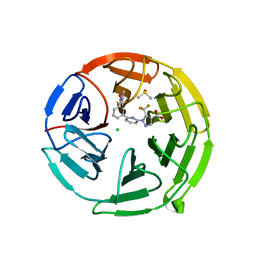 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | Descriptor: | 1-[6-[3-(dimethylcarbamoyl)phenyl]pyridin-2-yl]-5-(trifluoromethyl)pyrazole-4-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Davies, T.G, Cleasby, A. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
6VQ2
 
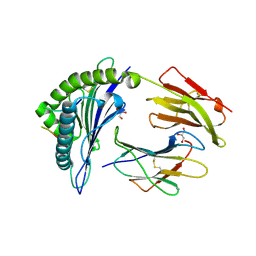 | | HLA-B*27:05 presenting an HIV-1 14mer peptide | | Descriptor: | 14-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
