2QUT
 
 | |
2R0D
 
 | | Crystal Structure of Autoinhibited Form of Grp1 Arf GTPase Exchange Factor | | Descriptor: | Cytohesin-3, DI(HYDROXYETHYL)ETHER, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ... | | Authors: | DiNitto, J.P, Delprato, A, Gabe Lee, M.T, Cronin, T.C, Huang, S, Guilherme, A, Czech, M.P, Lambright, D.G. | | Deposit date: | 2007-08-18 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis and Mechanism of Autoregulation in 3-Phosphoinositide-Dependent Grp1 Family Arf GTPase Exchange Factors.
Mol.Cell, 28, 2007
|
|
2BBN
 
 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | Descriptor: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | Authors: | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | Deposit date: | 1992-07-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
2RAP
 
 | | THE SMALL G PROTEIN RAP2A IN COMPLEX WITH GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RAP2A | | Authors: | Cherfils, J, Menetrey, J, Lebras, G. | | Deposit date: | 1998-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the small G protein Rap2A in complex with its substrate GTP, with GDP and with GTPgammaS.
EMBO J., 16, 1997
|
|
2P2T
 
 | | Crystal structure of dynein light chain LC8 bound to residues 123-138 of intermediate chain IC74 | | Descriptor: | ACETATE ION, Dynein intermediate chain peptide, Dynein light chain 1, ... | | Authors: | Benison, G, Karplus, P.A, Barbar, E. | | Deposit date: | 2007-03-07 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and dynamics of LC8 complexes with KXTQT-motif peptides: swallow and dynein intermediate chain compete for a common site.
J.Mol.Biol., 371, 2007
|
|
4JZ4
 
 | | Crystal structure of chicken c-Src-SH3 domain: monomeric form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Camara-Artigas, A. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Electrostatic Effects in the Folding of the SH3 Domain of the c-Src Tyrosine Kinase: pH-Dependence in 3D-Domain Swapping and Amyloid Formation.
Plos One, 9, 2014
|
|
2BID
 
 | | HUMAN PRO-APOPTOTIC PROTEIN BID | | Descriptor: | PROTEIN (BID) | | Authors: | Chou, J.J, Li, H, Salvesen, G.S, Yuan, J, Wagner, G. | | Deposit date: | 1999-01-27 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BID, an intracellular amplifier of apoptotic signaling.
Cell(Cambridge,Mass.), 96, 1999
|
|
4JZ3
 
 | | Crystal structure of the chicken c-Src-SH3 domain intertwined dimer | | Descriptor: | DI(HYDROXYETHYL)ETHER, Proto-oncogene tyrosine-protein kinase Src, TRIETHYLENE GLYCOL | | Authors: | Camara-Artigas, A. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Electrostatic Effects in the Folding of the SH3 Domain of the c-Src Tyrosine Kinase: pH-Dependence in 3D-Domain Swapping and Amyloid Formation.
Plos One, 9, 2014
|
|
2C47
 
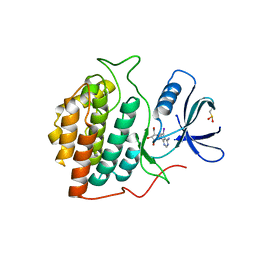 | | Structure of casein kinase 1 gamma 2 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, CASEIN KINASE 1 GAMMA 2 ISOFORM, MAGNESIUM ION | | Authors: | Bunkoczi, G, Rellos, P, Das, S, Ugochukwu, E, Fedorov, O, Sobott, F, Eswaran, J, Amos, A, Ball, L, von Delft, F, Bullock, A, Debreczeni, J, Turnbull, A, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-10-16 | | Release date: | 2005-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of Casein Kinase 1 Gamma 2
To be Published
|
|
2MBG
 
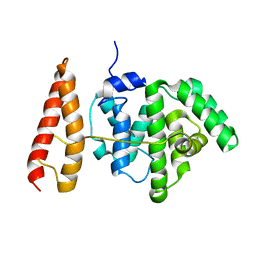 | | Rlip76 (gap-gbd) | | Descriptor: | RalA-binding protein 1 | | Authors: | Rajasekar, K.V, Campbell, L.J, Nietlispach, D, Owen, D, Mott, H.R. | | Deposit date: | 2013-07-30 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of the RLIP76 RhoGAP-Ral Binding Domain Dyad: Fixed Position of the Domains Leads to Dual Engagement of Small G Proteins at the Membrane.
Structure, 21, 2013
|
|
1SRL
 
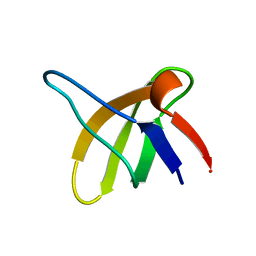 | | 1H AND 15N ASSIGNMENTS AND SECONDARY STRUCTURE OF THE SRC SH3 DOMAIN | | Descriptor: | SRC TYROSINE KINASE SH3 DOMAIN | | Authors: | Yu, H, Rosen, M.K, Shin, T.B, Seidel-Dugan, C, Brugge, J.S, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N assignments and secondary structure of the Src SH3 domain.
FEBS Lett., 324, 1993
|
|
1SDF
 
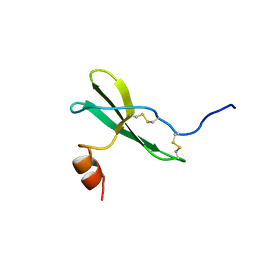 | | SOLUTION STRUCTURE OF STROMAL CELL-DERIVED FACTOR-1 (SDF-1), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STROMAL CELL-DERIVED FACTOR-1 | | Authors: | Crump, M.P, Rajarathnam, K, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 1997-11-15 | | Release date: | 1998-01-28 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1.
EMBO J., 16, 1997
|
|
1OOT
 
 | |
2M9N
 
 | | Solution Structure of (HhH)2 domain of human FAAP24 | | Descriptor: | Fanconi anemia-associated protein of 24 kDa | | Authors: | Wu, F, Han, X, Shi, C, Gong, W, Tian, C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure analysis of FAAP24 reveals single-stranded DNA-binding activity and domain functions in DNA damage response.
Cell Res., 23, 2013
|
|
1U81
 
 | | Delta-17 Human ADP Ribosylation Factor 1 Complexed with GDP | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Seidel, R.D, Amor, J.C, Kahn, R.A, Prestegard, J.H. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational changes in human Arf1 on nucleotide exchange and deletion of membrane-binding elements.
J.Biol.Chem., 279, 2004
|
|
3VOQ
 
 | |
1PRM
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1PRL
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
5L9B
 
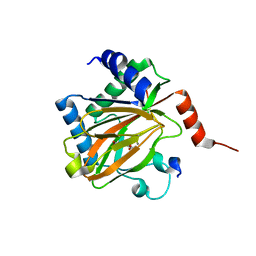 | |
5LA9
 
 | |
2MUU
 
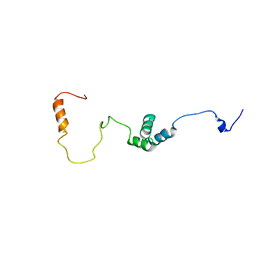 | |
2DNP
 
 | | Solution structure of RNA binding domain 2 in RNA-binding protein 14 | | Descriptor: | RNA-binding protein 14 | | Authors: | Kusuhara, M, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain 2 in RNA-binding protein 14
To be Published
|
|
1TUU
 
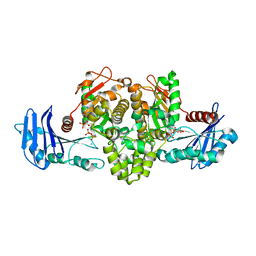 | | Acetate Kinase crystallized with ATPgS | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, AMMONIUM ION, ... | | Authors: | Gorrell, A, Lawrence, S.H, Ferry, J.G. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and kinetic analyses of arginine residues in the active site of the acetate kinase from Methanosarcina thermophila.
J. Biol. Chem., 280, 2005
|
|
4D2G
 
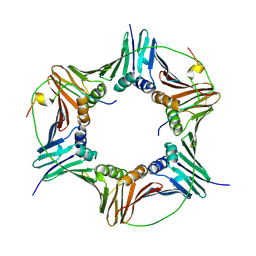 | | Crystal structure of human PCNA in complex with p15 peptide | | Descriptor: | P15, PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | DeBiasio, A, Ibanez, A, Mortuza, G, Molina, R, Cordeiro, T.N, Castillo, F, Villate, M, Merino, N, Lelli, M, Diercks, T, Luque, I, Bernardo, P, Montoya, G, Blanco, F.J. | | Deposit date: | 2014-05-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of P15(Paf)-PCNA Complex and Implications for Clamp Sliding During DNA Replication and Repair.
Nat.Commun., 6, 2015
|
|
1A15
 
 | | SDF-1ALPHA | | Descriptor: | STROMAL DERIVED FACTOR-1ALPHA, SULFATE ION | | Authors: | Dealwis, C.G, Fernandez, E.J, Lolis, E. | | Deposit date: | 1997-12-22 | | Release date: | 1998-08-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of chemically synthesized [N33A] stromal cell-derived factor 1alpha, a potent ligand for the HIV-1 "fusin" coreceptor.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
