6JQE
 
 | | The structural basis of the beta-carbonic anhydrases CafD (wild type) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-03-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of the low catalytic activities of the two minor beta-carbonic anhydrases of the filamentous fungus Aspergillus fumigatus.
J.Struct.Biol., 208, 2019
|
|
6VD8
 
 | |
6UXM
 
 | |
6UXU
 
 | |
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | Descriptor: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | Deposit date: | 2015-10-07 | | Release date: | 2016-03-16 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
6V74
 
 | | Crystal Structure of Human PKM2 in Complex with L-asparagine | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ASPARAGINE, CHLORIDE ION, ... | | Authors: | Nandi, S, Dey, M. | | Deposit date: | 2019-12-07 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Biochemical and structural insights into how amino acids regulate pyruvate kinase muscle isoform 2.
J.Biol.Chem., 295, 2020
|
|
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | Descriptor: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | Deposit date: | 2019-11-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
1ANP
 
 | |
6JQC
 
 | | The structural basis of the beta-carbonic anhydrase CafC (wild type) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-03-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of the low catalytic activities of the two minor beta-carbonic anhydrases of the filamentous fungus Aspergillus fumigatus.
J.Struct.Biol., 208, 2019
|
|
1A4Y
 
 | | RIBONUCLEASE INHIBITOR-ANGIOGENIN COMPLEX | | Descriptor: | ANGIOGENIN, RIBONUCLEASE INHIBITOR | | Authors: | Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 1998-02-08 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular recognition of human angiogenin by placental ribonuclease inhibitor--an X-ray crystallographic study at 2.0 A resolution.
EMBO J., 16, 1997
|
|
6JQD
 
 | | The structural basis of the beta-carbonic anhydrase CafC (L25G and L78G mutant) of the filamentous fungus Aspergillus fumigatus | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Jin, M.S, Kim, S, Kim, N.J, Hong, S, Kim, S, Sung, J. | | Deposit date: | 2019-03-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The structural basis of the low catalytic activities of the two minor beta-carbonic anhydrases of the filamentous fungus Aspergillus fumigatus.
J.Struct.Biol., 208, 2019
|
|
3PVN
 
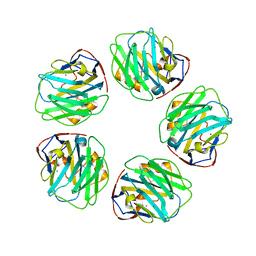 | | Triclinic form of Human C-Reactive Protein in complex with Zinc | | Descriptor: | C-reactive protein, CALCIUM ION, ZINC ION | | Authors: | Guillon, C, Mavoungou Bigouagou, U, Jeannin, P, Delneste, Y, Gouet, P. | | Deposit date: | 2010-12-07 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Staggered Decameric Assembly of Human C-Reactive Protein Stabilized by Zinc Ions Revealed by X-ray Crystallography.
Protein Pept.Lett., 22, 2014
|
|
1BXG
 
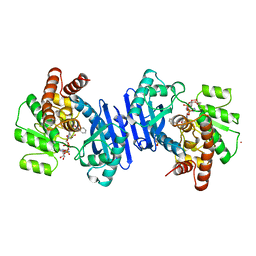 | | PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND BETA-PHENYLPROPIONATE | | Descriptor: | HYDROCINNAMIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B, Brunhuber, N.M.W, Blanchard, J.L, Holden, H.M. | | Deposit date: | 1998-10-02 | | Release date: | 1999-05-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenylalanine dehydrogenase from Rhodococcus sp. M4: high-resolution X-ray analyses of inhibitory ternary complexes reveal key features in the oxidative deamination mechanism.
Biochemistry, 38, 1999
|
|
1BSR
 
 | |
1ZBL
 
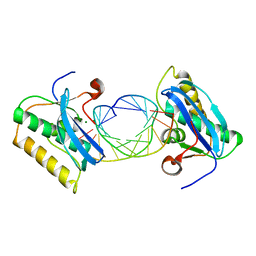 | | Bacillus halodurans RNase H catalytic domain mutant D192N in complex with 12-mer RNA/DNA hybrid | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3', 5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3', MAGNESIUM ION, ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis.
Cell(Cambridge,Mass.), 121, 2005
|
|
6WGI
 
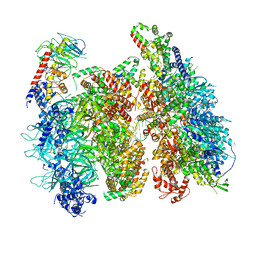 | | Atomic model of the mutant OCCM (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) loaded on DNA at 10.5 A resolution | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (34-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6HIL
 
 | |
6TMO
 
 | | Structure determination of an enhanced affinity TCR, a24b17, in complex with HLA-A*02:01 presenting a MART-1 peptide, EAAGIGILTV | | Descriptor: | 1,2-ETHANEDIOL, Alpha chain of engineered high affinity T-cell receptor, Beta chain of high affinity engineered T-cell receptor, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Rules Underpinning Enhanced Affinity Binding of Human T Cell Receptors Engineered for Immunotherapy.
Mol Ther Oncolytics, 18, 2020
|
|
6TNF
 
 | | Structure of monoubiquitinated FANCD2 in complex with FANCI and DNA | | Descriptor: | DNA (33-MER), FANCD2, Fanconi anemia complementation group I, ... | | Authors: | Alcon, P, Shakeel, S, Passmore, L.A. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6TQA
 
 | | X-ray structure of Roquin ROQ domain in complex with a UCP3 CDE2 SL RNA motif | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA (5'-R(P*GP*GP*UP*GP*CP*CP*UP*AP*AP*UP*AP*UP*UP*UP*AP*GP*GP*CP*AP*CP*(CCC))-3'), ... | | Authors: | Binas, O, Tants, J.-N, Peter, S.A, Janowski, R, Davydova, E, Braun, J, Niessing, D, Schwalbe, H, Weigand, J.E, Schlundt, A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the recognition of transiently structured AU-rich elements by Roquin.
Nucleic Acids Res., 48, 2020
|
|
2GF5
 
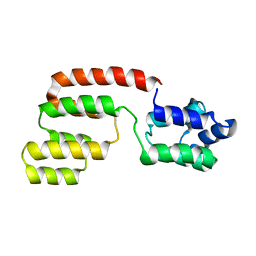 | | Structure of intact FADD (MORT1) | | Descriptor: | FADD protein | | Authors: | Carrington, P.E, Sandu, C, Wei, Y, Hill, J.M, Morisawa, G, Huang, T, Gavathiotis, E, Wei, Y, Werner, M.H. | | Deposit date: | 2006-03-21 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of FADD and Its Mode of Interaction with Procaspase-8
Mol.Cell, 22, 2006
|
|
6VD9
 
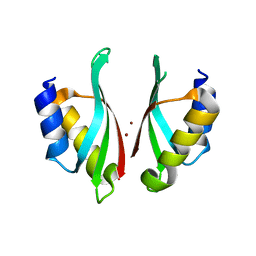 | |
6V89
 
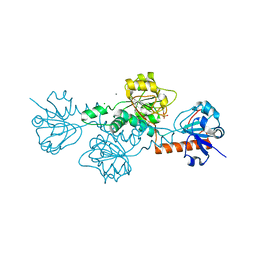 | | Human CtBP1 (28-375) in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, C-terminal-binding protein 1, CALCIUM ION, ... | | Authors: | Royer, W.E. | | Deposit date: | 2019-12-10 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | NAD(H) phosphates mediate tetramer assembly of human C-terminal binding protein (CtBP).
J.Biol.Chem., 296, 2021
|
|
8R79
 
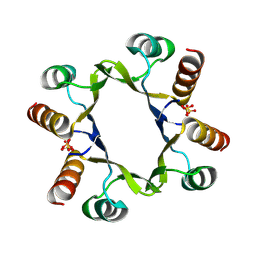 | | The D2 domain of human DTX3L | | Descriptor: | E3 ubiquitin-protein ligase DTX3L, SULFATE ION | | Authors: | Vela-Rodriguez, C, Lehtio, L, Maksimainen, M, Duman, R, Wagner, A, Glumoff, T. | | Deposit date: | 2023-11-24 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Oligomerisation mediated by the D2 domain of DTX3L is critical for DTX3L-PARP9 reading function of mono-ADP-ribosylated androgen receptor.
Biorxiv, 2023
|
|
6V3R
 
 | | Crystal structure of murine cycloxygenase in complex with a harmaline analog, 4,9-dihydro-3H-pyrido[3,4-b]indole | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-[(4-chlorophenyl)methyl]-6-methoxy-1-methyl-4,9-dihydro-3H-beta-carboline, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2019-11-26 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Harmaline Analogs as Substrate-Selective Cyclooxygenase-2 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
