4KRL
 
 | |
4K84
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 16:0 ceramide-1-phosphate (16:0-C1P) | | Descriptor: | (2S,3R,4E)-2-(hexadecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-17 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4K8E
 
 | | OYE1-W116V complexed with the aromatic product of (R)-carvone dismutation | | Descriptor: | 2-methyl-5-(prop-1-en-2-yl)phenol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sullivan, B, Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the
Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
4ZAY
 
 | | Structure of UbiX E49Q in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
4KT8
 
 | | The complex structure of Rv3378c-Y51FY90F with substrate, TPP | | Descriptor: | (2E)-3-methyl-5-[(1R,2S,8aS)-1,2,5,5-tetramethyl-1,2,3,5,6,7,8,8a-octahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
4ZAZ
 
 | | Structure of UbiX Y169F in complex with a covalent adduct formed between reduced FMN and dimethylallyl monophosphate | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, SODIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
4KRM
 
 | |
4ZI7
 
 | | CRYSTAL STRUCTURE OF TUBULIN-STATHMIN-TTL-HTI286 COMPLEX | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-04-27 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
4ZQN
 
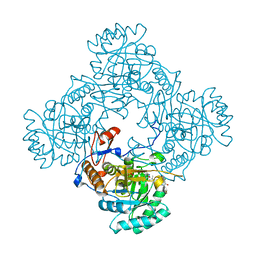 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor P41 | | Descriptor: | 2-chloro-N,N-dimethyl-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]benzamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4KRO
 
 | |
4KOY
 
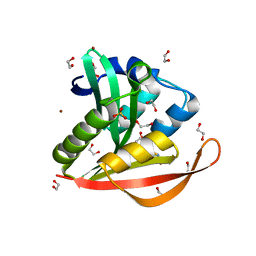 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cephalosporin C | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, SULFATE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KF6
 
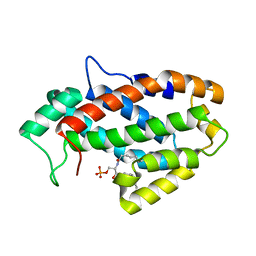 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 8:0 Ceramide-1-Phosphate (8:0-C1P) | | Descriptor: | (2S,3R,4E)-3-hydroxy-2-(octanoylamino)octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4ZAV
 
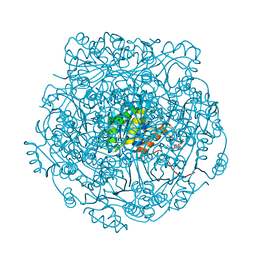 | | UbiX in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, SODIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
4K85
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 12:0 Ceramide-1-Phosphate (12:0-C1P) | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-17 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4K8N
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 18:1 Ceramide-1-Phosphate (18:1-C1P) | | Descriptor: | (2S,3R,4Z)-3-hydroxy-2-[(9E)-octadec-9-enoylamino]octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-18 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4KRN
 
 | | Nanobody/VHH domain EgA1 | | Descriptor: | Nanobody/VHH domain EgA1, SULFATE ION | | Authors: | Ferguson, K.M, Schmitz, K.R. | | Deposit date: | 2013-05-16 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Structural Evaluation of EGFR Inhibition Mechanisms for Nanobodies/VHH Domains.
Structure, 21, 2013
|
|
4KO0
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH an anilinylpyrimidine derivative (JLJ-135) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-methoxypyrimidin-2-yl)amino]-2-[(3-methylbut-2-en-1-yl)oxy]benzonitrile, HIV-1 reverse transcriptase, ... | | Authors: | Das, K, Bauman, J.D, Arnold, E. | | Deposit date: | 2013-05-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Extension into the entrance channel of HIV-1 reverse transcriptase-Crystallography and enhanced solubility.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4ZJS
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia Californica (Ac-AChBP) containing the main immunogenic region (MIR) from the human alpha 1 subunit of the muscle nicotinic acetylcholine receptor in complex with anatoxin-A. | | Descriptor: | 1-[(1R,6R)-9-azabicyclo[4.2.1]non-2-en-2-yl]ethanone, Acetylcholine receptor subunit alpha,Soluble acetylcholine receptor,Acetylcholine receptor subunit alpha,Soluble acetylcholine receptor | | Authors: | Talley, T.T, Bobango, J, Wu, J, Park, J.F, Luo, J, Lindsatrom, J, Taylor, P. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2301 Å) | | Cite: | Main immunogenic region structure promotes binding of conformation-dependent myasthenia gravis autoantibodies, nicotinic acetylcholine receptor conformation maturation, and agonist sensitivity.
J. Neurosci., 29, 2009
|
|
4ZQP
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor MAD1 | | Descriptor: | 5'-O-({1-[(2E)-4-(4-hydroxy-6-methoxy-7-methyl-3-oxo-1,3-dihydro-2-benzofuran-5-yl)-2-methylbut-2-en-1-yl]-1H-1,2,3-triazol-4-yl}methyl)adenosine, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4K80
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 2:0 ceramide-1-phosphate (2:0-C1P) | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-17 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4J6B
 
 | |
4IFG
 
 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, Calmodulin-domain protein kinase 1, UNKNOWN ATOM OR ION | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, K, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
To be Published
|
|
4IMO
 
 | | Crystal structure of wild type human Lipocalin PGDS in complex with substrate analog U44069 | | Descriptor: | (5E)-7-{(1R,4S,5S,6R)-5-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-6-yl}hept-5-enoic acid, Lipocalin-type prostaglandin-D synthase, THIOCYANATE ION | | Authors: | Lim, S.M, Chen, D, Teo, H, Roos, A, Nyman, T, Tresaugues, L, Pervushin, K, Nordlund, P. | | Deposit date: | 2013-01-03 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and dynamic insights into substrate binding and catalysis of human lipocalin prostaglandin D synthase.
J.Lipid Res., 54, 2013
|
|
4K55
 
 | |
4KIO
 
 | | Kinase domain mutant of human Itk in complex with a covalently-binding inhibitor | | Descriptor: | 1-[(3S)-3-{[4-(morpholin-4-ylmethyl)-6-([1,3]thiazolo[5,4-b]pyridin-2-ylamino)pyrimidin-2-yl]amino}pyrrolidin-1-yl]prop-2-en-1-one, 1-[(3S)-3-{[4-(morpholin-4-ylmethyl)-6-([1,3]thiazolo[5,4-b]pyridin-2-ylamino)pyrimidin-2-yl]amino}pyrrolidin-1-yl]propan-1-one, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2013-05-02 | | Release date: | 2013-08-21 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of novel irreversible inhibitors of interleukin (IL)-2-inducible tyrosine kinase (Itk) by targeting cysteine 442 in the ATP pocket.
J.Biol.Chem., 288, 2013
|
|
