4QUA
 
 | | Caspase-3 Y195F | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, Caspase-3 | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
4QUB
 
 | | Caspase-3 K137A | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, AZIDE ION, Caspase-3 | | Authors: | Cade, C, Swartz, P.D, MacKenzie, S.H, Clark, A.C. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Modifying caspase-3 activity by altering allosteric networks.
Biochemistry, 53, 2014
|
|
4JR0
 
 | | Human procaspase-3 bound to Ac-DEVD-CMK | | Descriptor: | Ac-DEVD-CMK, CHLORIDE ION, Procaspase-3 | | Authors: | Thomsen, N.D, Wells, J.A. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JQY
 
 | | Human procaspase-3, crystal form 1 | | Descriptor: | Procaspase-3 | | Authors: | Thomsen, N.D, Wells, J.A. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ITN
 
 | | Crystal structure of Pseudo-activated Procaspase-3 | | Descriptor: | ACETYL-ASP-GLU-VAL-ASP-CHLOROMETHYL KETONE inhibitor, Caspase-3 | | Authors: | Walters, J, Pop, C, Scott, F.L, Drag, M, Swartz, P.D, Mattos, C, Salvesen, G.S, Clark, A.C. | | Deposit date: | 2009-08-28 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A constitutively active and uninhibitable caspase-3 zymogen efficiently induces apoptosis.
Biochem.J., 424, 2009
|
|
5Y0Y
 
 | | RVFV GN-AU | | Descriptor: | GOLD ION, NSmGnGc | | Authors: | Wu, Y, Gao, F, Qi, J.X, Chai, Y, Gao, G.F. | | Deposit date: | 2017-07-19 | | Release date: | 2017-09-13 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3KJF
 
 | | Caspase 3 Bound to a covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,10aS)-2-{(2S)-4-carboxy-2-[(phenylacetyl)amino]butyl}-1,3-dioxo-2,3,5,7,8,9,10,10a-octahydro-1H-[1,2,4]triazolo[1,2-a]cinnolin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-3 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
5Y0W
 
 | | The structure of RVFV Gn head domain | | Descriptor: | NSmGnGc | | Authors: | Wu, Y, Gao, F, Qi, J.X, Chai, Y, Gao, G.F. | | Deposit date: | 2017-07-19 | | Release date: | 2017-09-13 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3DEI
 
 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | (1S)-2-oxo-1-phenyl-2-[(1,3,4-trioxo-1,2,3,4-tetrahydroisoquinolin-5-yl)amino]ethyl acetate, Caspase-3 | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
3DEK
 
 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | Caspase-3, N-[3-(2-fluoroethoxy)phenyl]-N'-(1,3,4-trioxo-1,2,3,4-tetrahydroisoquinolin-6-yl)butanediamide | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
2J30
 
 | | The Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)caspase-3 | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CASPASE-3 | | Authors: | Feeney, B, Pop, C, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)Caspase-3.
Biochemistry, 45, 2006
|
|
2J33
 
 | | The Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)caspase-3 | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CASPASE-3 | | Authors: | Feeney, B, Pop, C, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)Caspase-3.
Biochemistry, 45, 2006
|
|
2J31
 
 | | The Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of(Pro)caspase-3 | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CASPASE-3 | | Authors: | Feeney, B, Pop, C, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)Caspase-3.
Biochemistry, 45, 2006
|
|
2H65
 
 | | Crystal strusture of caspase-3 with inhibitor Ac-VDVAD-Cho | | Descriptor: | Ac-VDVAD-Cho, caspase-3, p12 subunit, ... | | Authors: | Fang, B, Boross, P.I, Tozser, J, Weber, I.T. | | Deposit date: | 2006-05-30 | | Release date: | 2006-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of caspase-3 reveals role for s5 binding site in substrate recognition
J.Mol.Biol., 360, 2006
|
|
3PCX
 
 | | Caspase-3 E246A, K242A Double Mutant | | Descriptor: | Caspase-3, Inhibitor Ac-DEVD-CMK | | Authors: | Walters, J, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2010-10-22 | | Release date: | 2011-02-23 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thermodynamic, enzymatic and structural effects of removing a salt bridge at the base of loop 4 in (pro)caspase-3.
Arch.Biochem.Biophys., 508, 2011
|
|
3PD1
 
 | | Caspase-3 K242A | | Descriptor: | Caspase-3, Inhibitor Ac-DEVD-CMK | | Authors: | Walters, J, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2010-10-22 | | Release date: | 2011-02-23 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Thermodynamic, enzymatic and structural effects of removing a salt bridge at the base of loop 4 in (pro)caspase-3.
Arch.Biochem.Biophys., 508, 2011
|
|
2J32
 
 | | The Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of(Pro)caspase-3 | | Descriptor: | ACE-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE INHIBITOR, CASPASE-3 | | Authors: | Feeney, B, Pop, C, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Role of Loop Bundle Hydrogen Bonds in the Maturation and Activity of (Pro)Caspase-3.
Biochemistry, 45, 2006
|
|
3PD0
 
 | | Caspase-3 E246A | | Descriptor: | CHLORIDE ION, Caspase-3, INHIBITOR AC-DEVD-CMK | | Authors: | Walters, J, Swartz, P, Mattos, C, Clark, A.C. | | Deposit date: | 2010-10-22 | | Release date: | 2011-02-16 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic, enzymatic and structural effects of removing a salt bridge at the base of loop 4 in (pro)caspase-3.
Arch.Biochem.Biophys., 508, 2011
|
|
3OD2
 
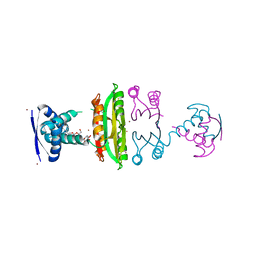 | | E. coli NikR soaked with excess nickel ions | | Descriptor: | 3-CYCLOHEXYLPROPYL 4-O-ALPHA-D-GLUCOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, NICKEL (II) ION, Nickel-responsive regulatory protein | | Authors: | Phillips, C.M, Schreiter, E.R, Drennan, C.L. | | Deposit date: | 2010-08-10 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of low-affinity nickel binding to the nickel-responsive transcription factor NikR from Escherichia coli.
Biochemistry, 49, 2010
|
|
7RNC
 
 | |
2KAE
 
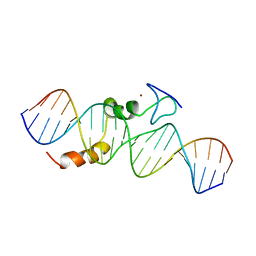 | | data-driven model of MED1:DNA complex | | Descriptor: | 5'-D(*DCP*DGP*DGP*DAP*DAP*DAP*DAP*DGP*DTP*DAP*DTP*DAP*DCP*DTP*DTP*DTP*DTP*DCP*DCP*DG)-3', GATA-type transcription factor, ZINC ION | | Authors: | Lowry, J.A, Gamsjaeger, R, Thong, S, Hung, W, Kwan, A.H, Broitman-Maduro, G, Matthews, J.M, Maduro, M, Mackay, J.P. | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of MED-1 Reveals Unexpected Diversity in the Mechanism of DNA Recognition by GATA-type Zinc Finger Domains.
J.Biol.Chem., 284, 2009
|
|
2H5I
 
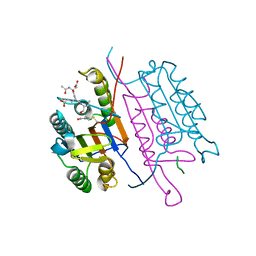 | | Crystal structure of caspase-3 with inhibitor Ac-DEVD-Cho | | Descriptor: | Ac-DEV(ASJ), caspase-3, p12 subunit, ... | | Authors: | Fang, B, Boross, P.I, Tozser, J, Weber, I.T. | | Deposit date: | 2006-05-26 | | Release date: | 2006-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and kinetic analysis of caspase-3 reveals role for s5 binding site in substrate recognition
J.Mol.Biol., 360, 2006
|
|
4MDC
 
 | | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021, NYSGRC target 021389 | | Descriptor: | GLYCEROL, Putative glutathione S-transferase | | Authors: | Shabalin, I.G, Bacal, P, Cooper, D.R, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-22 | | Release date: | 2013-09-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021
To be Published
|
|
2H5J
 
 | | Crystal strusture of caspase-3 with inhibitor Ac-DMQD-Cho | | Descriptor: | Ac-DMQD-Cho, caspase-3, p12 subunit, ... | | Authors: | Fang, B, Boross, P.I, Tozser, J, Weber, I.T. | | Deposit date: | 2006-05-26 | | Release date: | 2006-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic analysis of caspase-3 reveals role for s5 binding site in substrate recognition
J.Mol.Biol., 360, 2006
|
|
1RHJ
 
 | |
