7M8K
 
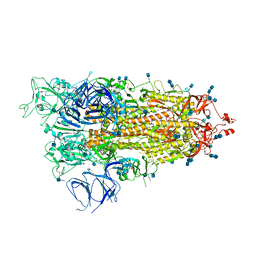 | | Cryo-EM structure of Brazil (P.1) SARS-CoV-2 spike glycoprotein variant in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L, Ho, D.D. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization.
Cell Host Microbe, 29, 2021
|
|
8CSG
 
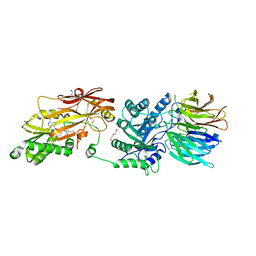 | | Human PRMT5:MEP50 structure with Fragment 1 and MTA Bound | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, 6-bromo-1H-pyrrolo[3,2-b]pyridin-5-amine, ... | | Authors: | Gunn, R.J, Lawson, J.D, Smith, C.R. | | Deposit date: | 2022-05-12 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
4NBU
 
 | | Crystal structure of FabG from Bacillus sp | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-(Acyl-carrier-protein) reductase, ACETOACETYL-COENZYME A | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
5LYM
 
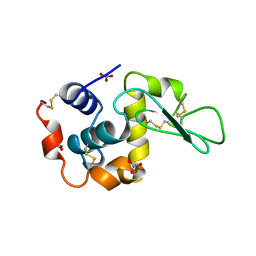 | |
5M6N
 
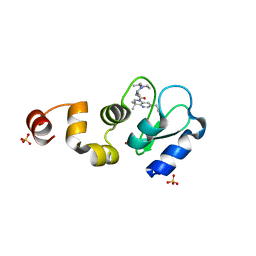 | | Small Molecule inhibitors of IAP | | Descriptor: | 1-[6-[(4-fluorophenyl)methyl]-3,3-dimethyl-2~{H}-pyrrolo[3,2-b]pyridin-1-yl]-2-[(2~{R},5~{R})-5-methyl-2-[[(3~{R})-3-methylmorpholin-4-yl]methyl]piperazin-4-ium-1-yl]ethanone, Baculoviral IAP repeat-containing protein 2, SULFATE ION, ... | | Authors: | Williams, P.A. | | Deposit date: | 2016-10-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Potent Nonpeptidomimetic, Small-Molecule Antagonist of Cellular Inhibitor of Apoptosis Protein 1 (cIAP1) and X-Linked Inhibitor of Apoptosis Protein (XIAP).
J. Med. Chem., 60, 2017
|
|
5M6H
 
 | | Small Molecule inhibitors of IAP | | Descriptor: | 1-[3,3-dimethyl-6-(phenylmethyl)-2~{H}-pyrrolo[3,2-b]pyridin-1-yl]-2-[(2~{R},5~{R})-5-methyl-2-morpholin-4-ylcarbonyl-piperazin-4-ium-1-yl]ethanone, E3 ubiquitin-protein ligase XIAP, SODIUM ION, ... | | Authors: | Williams, P.A. | | Deposit date: | 2016-10-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a Potent Nonpeptidomimetic, Small-Molecule Antagonist of Cellular Inhibitor of Apoptosis Protein 1 (cIAP1) and X-Linked Inhibitor of Apoptosis Protein (XIAP).
J. Med. Chem., 60, 2017
|
|
6SYI
 
 | |
5M6F
 
 | | Small Molecule inhibitors of IAP | | Descriptor: | 1-[3,3-dimethyl-6-(phenylmethyl)-2~{H}-pyrrolo[3,2-b]pyridin-1-yl]-2-[(2~{R},5~{R})-2-(methoxymethyl)-5-methyl-piperazin-4-ium-1-yl]ethanone, E3 ubiquitin-protein ligase XIAP, SODIUM ION, ... | | Authors: | Williams, P.A. | | Deposit date: | 2016-10-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of a Potent Nonpeptidomimetic, Small-Molecule Antagonist of Cellular Inhibitor of Apoptosis Protein 1 (cIAP1) and X-Linked Inhibitor of Apoptosis Protein (XIAP).
J. Med. Chem., 60, 2017
|
|
7B3Q
 
 | | Crystal structure of c-MET bound by compound 1 | | Descriptor: | 1-(phenylmethyl)-5~{H}-pyrrolo[3,2-c]pyridin-4-one, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
6SHY
 
 | |
5FH8
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with compound 28 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranyl-3-(2-ethylbutyl)-4~{H}-pyrrolo[1,2-a]quinazolin-5-one, DIMETHYL SULFOXIDE, ... | | Authors: | Tallant, C, Sutherell, C.L, Siejka, P, Sorrell, F.J, Krojer, T, Picaud, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Ley, S.V, Knapp, S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Development of 2,3-Dihydropyrrolo[1,2-a]quinazolin-5(1H)-one Inhibitors Targeting Bromodomains within the Switch/Sucrose Nonfermenting Complex.
J.Med.Chem., 59, 2016
|
|
3TUB
 
 | |
5FMA
 
 | | human Notch 1, EGF 4-7 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Weisshuhn, P.C, Sheppard, D, Taylor, P, Whiteman, P, Lea, S.M, Handford, P.A, Redfield, C. | | Deposit date: | 2015-11-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Non-Linear and Flexible Regions of the Human Notch1 Extracellular Domain Revealed by High-Resolution Structural Studies.
Structure, 24, 2016
|
|
5FRO
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ446 (compound 4f) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-[2-(phenylsulfonyl)ethyl]-1,8-naphthyridine-3-carboxamide, 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*CP*GP*CP*A)-3', 5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP *AP*CP*A)-3', ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2015-12-18 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases.
Acs Chem.Biol., 11, 2016
|
|
7O4C
 
 | |
7O7I
 
 | |
6SMH
 
 | | Cryo-electron microscopy structure of a RbcL-Raf1 supercomplex from Synechococcus elongatus PCC 7942 | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Rubisco accumulation factor 1 (RAF1) peptide | | Authors: | Huang, F, Kong, W.-W, Sun, Y, Chen, T, Dykes, G.F, Jiang, Y.L, Liu, L.N. | | Deposit date: | 2019-08-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rubisco accumulation factor 1 (Raf1) plays essential roles in mediating Rubisco assembly and carboxysome biogenesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7O6X
 
 | | Tankyrase 2 in complex with an inhibitor (OM-153) | | Descriptor: | N-[3-[5-(5-ethoxypyridin-2-yl)-4-(2-fluorophenyl)-1,2,4-triazol-3-yl]cyclobutyl]quinoxaline-5-carboxamide, Poly [ADP-ribose] polymerase tankyrase-2, ZINC ION | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2021-04-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a 1,2,4-Triazole-Based Lead Tankyrase Inhibitor: Part II.
J.Med.Chem., 64, 2021
|
|
5FRI
 
 | | ALK5 in complex witha an N-(4-anilino-2-pyridyl)acetamide inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, N-[4-[(6-chloro-[1,3]dioxolo[4,5-b]pyridin-7-yl)amino]-2-pyridyl]cyclopropanecarboxamide, TGF-BETA RECEPTOR TYPE-1 | | Authors: | Goldberg, F.W, Daunt, P, Pearson, S.E, Greenwood, R, Debreczeni, J, Grist, M. | | Deposit date: | 2015-12-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and optimisation of a series of N-(4-anilino-2-pyridyl)acetamide activin receptor-like kinase 1 (ALK1) inhibitors
Med. Chem. Commun., 7, 2016
|
|
8CKW
 
 | |
8CL1
 
 | |
8CKV
 
 | |
8CL3
 
 | |
8CL4
 
 | |
8CKZ
 
 | |
