3IMS
 
 | | Transthyretin in complex with 2,6-dibromo-4-(2,6-dichlorophenethyl)phenol | | Descriptor: | 2,6-dibromo-4-[2-(2,6-dichlorophenyl)ethyl]phenol, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity.
J.Am.Chem.Soc., 132, 2010
|
|
1M4X
 
 | | PBCV-1 virus capsid, quasi-atomic model | | Descriptor: | PBCV-1 virus capsid | | Authors: | Nandhagopal, N, Simpson, A.A, Gurnon, J.R, Yan, X, Baker, T.S, Graves, M.V, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2002-07-05 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | The Structure and Evolution of the Major Capsid Protein of a Large,
Lipid containing, DNA virus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2J9N
 
 | | Robotically harvested Trypsin complexed with Benzamidine containing polypeptide mediated crystal contacts | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Viola, R, Carman, P, Walsh, J, Miller, E, Benning, M, Frankel, D, McPherson, A, Cudney, R, Rupp, B. | | Deposit date: | 2006-11-14 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Operator Assisted Harvesting of Protein Crystals Using a Universal Micromanipulation Robot.
J.Appl.Crystallogr., 40, 2007
|
|
6HZX
 
 | | Protein-aromatic foldamer complex crystal structure | | Descriptor: | Aromatic foldamer, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Post, S, Langlois d'Estaintot, B, Fischer, L, Granier, T, Huc, I. | | Deposit date: | 2018-10-24 | | Release date: | 2019-09-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure Elucidation of Helical Aromatic Foldamer-Protein Complexes with Large Contact Surface Areas.
Chemistry, 25, 2019
|
|
4X9S
 
 | | CRYSTAL STRUCTURE OF HISAP FROM STREPTOMYCES SP. MG1 | | Descriptor: | Phosphoribosyl isomerase A, SULFATE ION | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
5HST
 
 | |
5HWL
 
 | | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 2, N,N'-(butane-1,4-diyl)bis{2-[2,3-dichloro-4-(2-methylidenebutanoyl)phenoxy]acetamide} | | Authors: | Zhang, X, Wei, J, Wu, S, Zhang, H.P, Luo, M, Yang, X.L, Liao, F, Wang, D.Q. | | Deposit date: | 2016-01-29 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human glutathione s-transferase Mu2 complexed with BDEA, monoclinic crystal form
To Be Published
|
|
1MUW
 
 | |
1SK7
 
 | | Structural Basis for Novel Delta-Regioselective Heme Oxygenation in the Opportunistic Pathogen Pseudomonas aeruginosa | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, hypothetical protein pa-HO | | Authors: | Friedman, J, Lad, L, Li, H, Wilks, A, Poulos, T.L. | | Deposit date: | 2004-03-04 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for novel delta-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa
Biochemistry, 43, 2004
|
|
3I6W
 
 | |
1MWE
 
 | |
3IMT
 
 | | Transthyretin in complex with (E)-4-(4-aminostyryl)-2,6-dibromophenol | | Descriptor: | 4-[(E)-2-(4-aminophenyl)ethenyl]-2,6-dibromophenol, Transthyretin | | Authors: | Connelly, S, Wilson, I. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity.
J.Am.Chem.Soc., 132, 2010
|
|
6L02
 
 | |
5B0X
 
 | | Crystal structure of the CK2a/benzoic acid derivative complex | | Descriptor: | 4-[2-[(4-methoxyphenyl)carbonylamino]-1,3-thiazol-5-yl]benzoic acid, Casein kinase II subunit alpha | | Authors: | Kinoshita, T, Nakanishi, I. | | Deposit date: | 2015-11-13 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-activity relationship study of 4-(thiazol-5-yl)benzoic acid derivatives as potent protein kinase CK2 inhibitors
Bioorg.Med.Chem., 24, 2016
|
|
2IDQ
 
 | | Structure of M98A mutant of amicyanin, Cu(II) | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
3VRF
 
 | | The crystal structure of hemoglobin from woolly mammoth in the carbonmonoxy forms | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, ... | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1NP9
 
 | |
1NT6
 
 | | F1-Gramicidin C In Sodium Dodecyl Sulfate Micelles (NMR) | | Descriptor: | GRAMICIDIN C | | Authors: | Townsley, L.E, Fletcher, T.G, Hinton, J.F. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-11 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Structure, Cation Binding, Transport, and Conductance of Gly15-Gramicidin a Incorporated Into Sds Micelles and Pc/Pg Vesicles.
Biochemistry, 42, 2003
|
|
1SBH
 
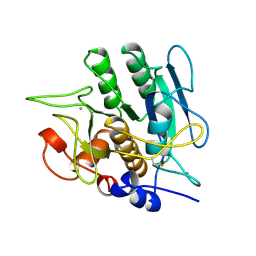 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) MUTANT (M50F, N76D, G169A, Q206C, N218S, K256Y) | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A weak calcium binding site in subtilisin BPN' has a dramatic effect on protein stability.
J.Am.Chem.Soc., 118, 1996
|
|
1SPB
 
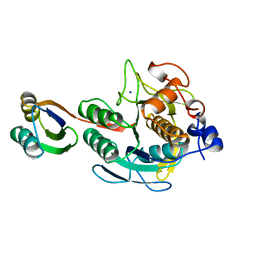 | | SUBTILISIN BPN' PROSEGMENT (77 RESIDUES) COMPLEXED WITH A MUTANT SUBTILISIN BPN' (266 RESIDUES). CRYSTAL PH 4.6. CRYSTALLIZATION TEMPERATURE 20 C DIFFRACTION TEMPERATURE-160 C | | Descriptor: | SODIUM ION, SUBTILISIN BPN', SUBTILISIN BPN' PROSEGMENT | | Authors: | Gallagher, D.T, Gilliland, G.L, Wang, L, Bryan, P.N. | | Deposit date: | 1995-06-21 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The prosegment-subtilisin BPN' complex: crystal structure of a specific 'foldase'.
Structure, 3, 1995
|
|
3IAL
 
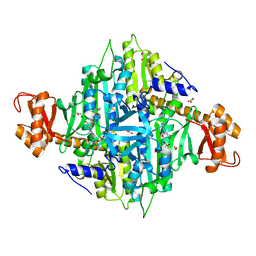 | | Giardia lamblia Prolyl-tRNA synthetase in complex with prolyl-adenylate | | Descriptor: | 5'-O-[(R)-hydroxy{[(2S)-pyrrolidin-2-ylcarbonyl]oxy}phosphoryl]adenosine, GLYCEROL, Prolyl-tRNA synthetase | | Authors: | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2009-07-14 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the prolyl-tRNA synthetase from the eukaryotic pathogen Giardia lamblia.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5AN8
 
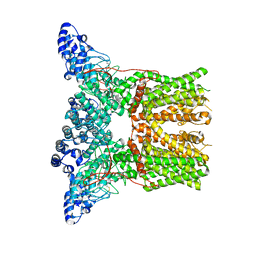 | | Cryo-electron microscopy structure of rabbit TRPV2 ion channel | | Descriptor: | TRPV2 | | Authors: | Zubcevic, L, Herzik, M.A.J, Chung, B.C, Lander, G.C, Lee, S.Y. | | Deposit date: | 2015-09-04 | | Release date: | 2015-12-23 | | Last modified: | 2019-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-Electron Microscopy of the Trpv2 Ion Channel
Nat.Struct.Mol.Biol., 23, 2016
|
|
6I8H
 
 | |
6IAM
 
 | |
3IFT
 
 | | Crystal structure of glycine cleavage system protein H from Mycobacterium tuberculosis, using X-rays from the Compact Light Source. | | Descriptor: | Glycine cleavage system H protein | | Authors: | Edwards, T.E, Abendroth, J, Staker, B, Mayer, C, Phan, I, Kelley, A, Analau, E, Leibly, D, Rifkin, J, Loewen, R, Ruth, R.D, Stewart, L.J, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2009-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of the glycine cleavage system protein H of Mycobacterium tuberculosis using an inverse Compton synchrotron X-ray source.
J.Struct.Funct.Genom., 11, 2010
|
|
