7RFS
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
6QZ3
 
 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | BENZOIC ACID, CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-11 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5LVP
 
 | | Human PDK1 Kinase Domain in Complex with an HM-Peptide Bound to the PIF-Pocket | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schulze, J.O, Saladino, G, Busschots, K, Neimanis, S, Suess, E, Odadzic, D, Zeuzem, S, Hindie, V, Herbrand, A.K, Lisa, M.N, Alzari, P.M, Gervasio, F.L, Biondi, R.M. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bidirectional Allosteric Communication between the ATP-Binding Site and the Regulatory PIF Pocket in PDK1 Protein Kinase.
Cell Chem Biol, 23, 2016
|
|
6BHJ
 
 | | Structure of HIV-1 Reverse Transcriptase Bound to a 38-mer Hairpin Template-Primer RNA-DNA Aptamer | | Descriptor: | 38-MER RNA-DNA Aptamer, GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Ruiz, F.X, Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Integrative Structural Biology Studies of HIV-1 Reverse Transcriptase Binding to a High-Affinity DNA Aptamer
Curr Res Struct Biol, 2020
|
|
8FUH
 
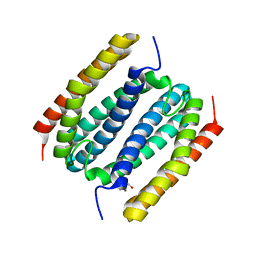 | |
6AVM
 
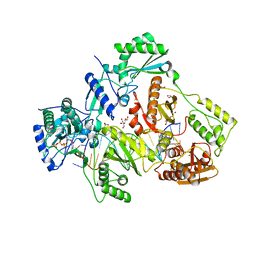 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 9.5 WITH CROSS-LINKING TO SECOND BASE TEMPLATE OVERHANG | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA (27-MER), DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DDG))-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-09-03 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
5KMG
 
 | | Near-atomic cryo-EM structure of PRC1 bound to the microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kellogg, E.H, Howes, S, Ti, S.-C, Ramirez-Aportela, E, Kapoor, T.M, Chacon, P, Nogales, E. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic cryo-EM structure of PRC1 bound to the microtubule.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1H24
 
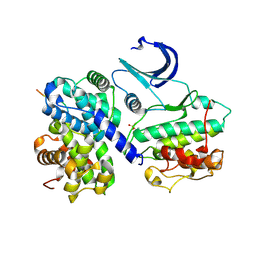 | | CDK2/CyclinA in complex with a 9 residue recruitment peptide from E2F | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, TRANSCRIPTION FACTOR E2F1 | | Authors: | Tews, I, Cheng, K.Y, Lowe, E.D, Noble, M.E.M, Brown, N.R, Gul, S, Gamblin, S, Johnson, L.N. | | Deposit date: | 2002-07-31 | | Release date: | 2003-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Specificity Determinants of Recruitment Peptides Bound to Phospho-Cdk2/Cyclin A
Biochemistry, 41, 2002
|
|
8EL2
 
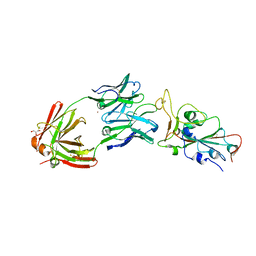 | | SARS-CoV-2 RBD bound to neutralizing antibody Fab ICO-hu23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ICO-hu23 Heavy Chain, Fab ICO-hu23 Light Chain, ... | | Authors: | Besaw, J.E, Kuo, A, Morizumi, T, Ernst, O.P. | | Deposit date: | 2022-09-22 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
5N9R
 
 | | Crystal structure of USP7 in complex with a potent, selective and reversible small-molecule inhibitor | | Descriptor: | 7-bromanyl-3-[[4-oxidanyl-1-[(3~{R})-3-phenylbutanoyl]piperidin-4-yl]methyl]thieno[3,2-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Harrison, T, Gavory, G, O'Dowd, C, Helm, M, Flasz, I, Arkoudis, E, Dossang, A, Hughes, C, Cassidy, E, McClelland, K, Odrzywol, E, Page, N, Barker, O, Miel, H. | | Deposit date: | 2017-02-27 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and characterization of highly potent and selective allosteric USP7 inhibitors.
Nat. Chem. Biol., 14, 2018
|
|
1H28
 
 | | CDK2/CyclinA in complex with an 11-residue recruitment peptide from p107 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, RETINOBLASTOMA-LIKE PROTEIN 1 | | Authors: | Tews, I, Cheng, K.Y, Lowe, E.D, Noble, M.E.M, Brown, N.R, Gul, S, Gamblin, S, Johnson, L.N. | | Deposit date: | 2002-07-31 | | Release date: | 2003-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specificity Determinants of Recruitment Peptides Bound to Phospho-Cdk2/Cyclin A
Biochemistry, 41, 2002
|
|
7UTJ
 
 | | Cryogenic electron microscopy 3D map of F-actin bound by human dimeric alpha-catenin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rangarajan, E.S, Smith, E.W, Izard, T. | | Deposit date: | 2022-04-27 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Distinct inter-domain interactions of dimeric versus monomeric alpha-catenin link cell junctions to filaments.
Commun Biol, 6, 2023
|
|
6AN8
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 8.0 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA PRIMER (5'- D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*GP)-3'), DNA TEMPLATE (5'- D(*AP*TP*GP*AP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
6OF8
 
 | | Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met mutant human CamKII-alpha hub domain | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha, GLYCEROL, POTASSIUM ION | | Authors: | McSpadden, E.D, Chi, C.C, Gee, C.L, Kuriyan, J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variation in assembly stoichiometry in non-metazoan homologs of the hub domain of Ca2+/calmodulin-dependent protein kinase II.
Protein Sci., 28, 2019
|
|
7U29
 
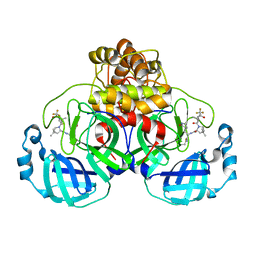 | | Structure of SARS-CoV-2 Mpro mutant (K90R) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2022-02-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
7U28
 
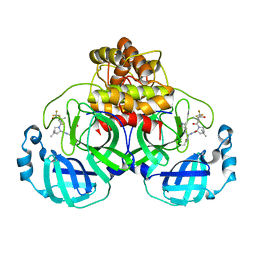 | | Structure of SARS-CoV-2 Mpro Lambda (G15S) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Greasley, S.E, Ferre, R.A, Plotnikova, O, Liu, W, Stewart, A.E. | | Deposit date: | 2022-02-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
1GY3
 
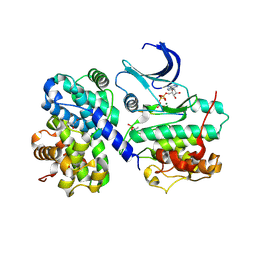 | | pCDK2/cyclin A in complex with MgADP, nitrate and peptide substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Cook, A, Lowe, E.D, Chrysina, E.D, Skamnaki, V.T, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 2002-04-19 | | Release date: | 2002-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Studies on Phospho-Cdk2/Cyclin a Bound to Nitrate, a Transition State Analogue: Implications for the Protein Kinase Mechanism
Biochemistry, 41, 2002
|
|
5IRV
 
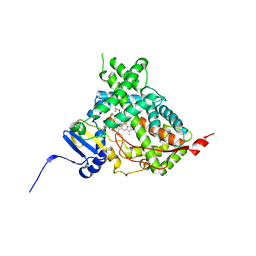 | | Human cytochrome P450 17A1 bound to inhibitor VT-464 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase, VT-464 | | Authors: | Scott, E.E, Petrunak, E.M. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Structural and Functional Evaluation of Clinically Relevant Inhibitors of Steroidogenic Cytochrome P450 17A1.
Drug Metab. Dispos., 45, 2017
|
|
8OF4
 
 | | Nucleosome Bound human SIRT6 (Composite) | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Smirnova, E, Bignon, E, Schultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Binding to nucleosome poises human SIRT6 for histone H3 deacetylation.
Elife, 12, 2024
|
|
1H27
 
 | | CDK2/CyclinA in complex with an 11-residue recruitment peptide from p27 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, CYCLIN-DEPENDENT KINASE INHIBITOR 1B | | Authors: | Tews, I, Cheng, K.Y, Lowe, E.D, Noble, M.E.M, Brown, N.R, Gul, S, Gamblin, S, Johnson, L.N. | | Deposit date: | 2002-07-31 | | Release date: | 2003-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Specificity Determinants of Recruitment Peptides Bound to Phospho-Cdk2/Cyclin A
Biochemistry, 41, 2002
|
|
1GSZ
 
 | | Crystal Structure of a Squalene Cyclase in Complex with the Potential Anticholesteremic Drug Ro48-8071 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE, [4-({6-[ALLYL(METHYL)AMINO]HEXYL}OXY)-2-FLUOROPHENYL](4-BROMOPHENYL)METHANONE | | Authors: | Lenhart, A, Weihofen, W.A, Pleschke, A.E.W, Schulz, G.E. | | Deposit date: | 2002-01-09 | | Release date: | 2003-01-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Squalene Cyclase in Complex with the Potential Anticholesteremic Drug Ro48-8071
Chem.Biol., 9, 2002
|
|
8QT5
 
 | |
6WYE
 
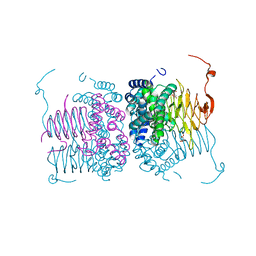 | | Crystal structure of Neisseria gonorrhoeae serine acetyltransferase (CysE) | | Descriptor: | (2S)-2-hydroxybutanedioic acid, SODIUM ION, Serine acetyltransferase | | Authors: | Hicks, J.L, Oldham, K.E, Summers, E.L, Prentice, E.J. | | Deposit date: | 2020-05-12 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Serine acetyltransferase from Neisseria gonorrhoeae; structural and biochemical basis of inhibition.
Biochem.J., 479, 2022
|
|
8UCU
 
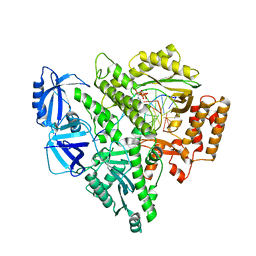 | | Partial DNA termination subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UCW
 
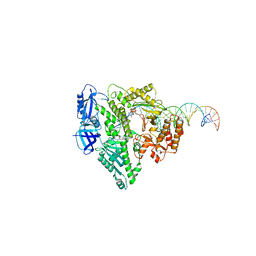 | | Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Chazin, W.C, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
