452D
 
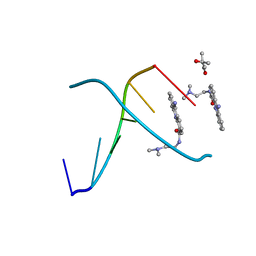 | | ACRIDINE BINDING TO DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Thorpe, J.H, Todd, A.K, Cardin, C.J. | | Deposit date: | 1999-02-18 | | Release date: | 2003-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Major groove binding and 'DNA-induced' fit in the intercalation of a derivative of the mixed topoisomerase I/II poison N-(2-(dimethlyamino)ethyl)acridine-4-carboxamide (DACA) into DNA: X-ray structure complexed to d(CG(5Br-U)ACG)2 at 1.3-angstrom resolution
J.Med.Chem., 42, 1999
|
|
1VS2
 
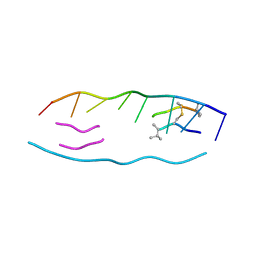 | | Interactions of quinoxaline antibiotic and DNA: the molecular structure of a TRIOSTIN A-D(GCGTACGC) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3', TRIOSTIN A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Rich, A. | | Deposit date: | 1986-10-21 | | Release date: | 2006-06-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions of Quinoxaline Antibiotic and DNA: The Molecular Structure of a Triostin A-D(Gcgtacgc) Complex.
J.Biomol.Struct.Dyn., 4, 1986
|
|
3HWU
 
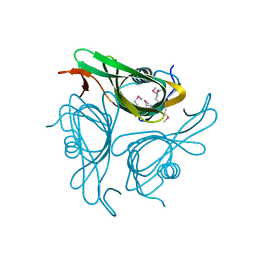 | |
6NFL
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G complexed with 2-HP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-diazinan-2-one, CHLORIDE ION, ... | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
6NFK
 
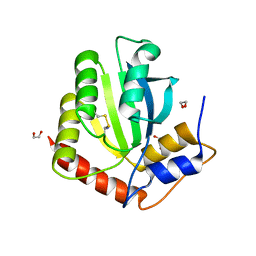 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G bound to iodide | | Descriptor: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IODIDE ION | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
4EFE
 
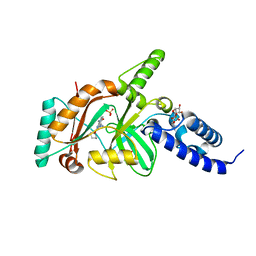 | | crystal structure of DNA ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of DNA ligase
To be Published
|
|
7DW5
 
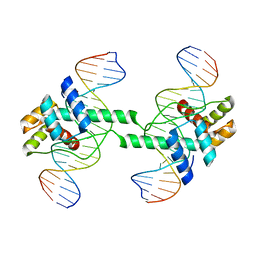 | | Crystal structure of DUX4 HD1-HD2 domain complexed with ERG sites | | Descriptor: | BROMIDE ION, DNA (5'-D(P*CP*GP*AP*CP*TP*TP*GP*AP*TP*GP*AP*GP*AP*TP*TP*AP*GP*AP*CP*TP*G)-3'), Double homeobox protein 4-like protein 2 | | Authors: | Zhang, H, Cheng, N, Li, Z, Zhang, W, Dong, X, Huang, J, Meng, G. | | Deposit date: | 2021-01-15 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | DNA crosslinking and recombination-activating genes 1/2 (RAG1/2) are required for oncogenic splicing in acute lymphoblastic leukemia.
Cancer Commun (Lond), 41, 2021
|
|
1VRR
 
 | | Crystal structure of the restriction endonuclease BstYI complex with DNA | | Descriptor: | 5'-D(*TP*TP*AP*TP*AP*GP*AP*TP*CP*TP*AP*TP*AP*A)-3', BstYI | | Authors: | Townson, S.A, Samuelson, J.C, Xu, S.Y, Aggarwal, A.K. | | Deposit date: | 2005-06-02 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Implications for Switching Restriction Enzyme Specificities from the Structure of BstYI Bound to a BglII DNA Sequence.
Structure, 13, 2005
|
|
2W9M
 
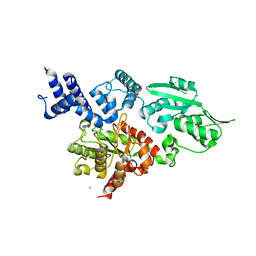 | | Structure of family X DNA polymerase from Deinococcus radiodurans | | Descriptor: | MERCURY (II) ION, POLYMERASE X, ZINC ION | | Authors: | Leulliot, N, Cladiere, L, Lecointe, F, Durand, D, Hubscher, U, van Tilbeurgh, H. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The Family X DNA Polymerase from Deinococcus Radioduran Adopts a Non-Standard Extended Conformation.
J.Biol.Chem., 284, 2009
|
|
3GO3
 
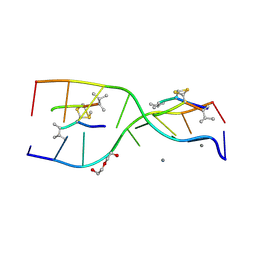 | | Interactions of an echinomycin-DNA complex with manganese(II) ions | | Descriptor: | 2-CARBOXYQUINOXALINE, 5'-D(*AP*CP*GP*TP*AP*CP*GP*T)-3', DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pfoh, R, Cuesta-Seijo, J.A, Sheldrick, G.M. | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Interaction of an Echinomycin-DNA Complex with Manganese Ion
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7Z3X
 
 | | Crystal structure of FIR RRM1-2 Y115F mutant bound to FUSE ssDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*T)-3'), DNA (5'-D(P*GP*TP*())-3'), ... | | Authors: | Ni, X, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of FIR RRM1-2 Y115F mutant bound to FUSE ssDNA
To Be Published
|
|
3N0U
 
 | |
2W58
 
 | | Crystal Structure of the DnaI | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, K.L, Lo, Y.H, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2008-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Interplay between the Replicative Helicase Dnac and its Loader Protein Dnai from Geobacillus Kaustophilus.
J.Mol.Biol., 393, 2009
|
|
4CXO
 
 | | bifunctional endonuclease in complex with ssDNA | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ENDONUCLEASE 2, SULFATE ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
4HQI
 
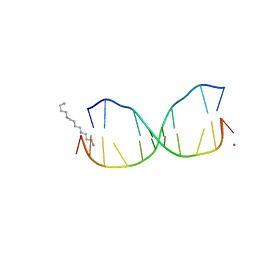 | | Structure of O6-Benzyl-2'-deoxyguanosine opposite perimidinone-derived synthetic nucleoside in DNA duplex | | Descriptor: | SPERMINE, STRONTIUM ION, Short modified nucleic acids | | Authors: | Kowal, E.A, Lad, R, Pallan, P.S, Muffly, E, Wawrzak, Z, Egli, M, Sturla, S.J, Stone, M.P. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of O6-benzyl-2'-deoxyguanosine by a perimidinone-derived synthetic nucleoside: a DNA interstrand stacking interaction.
Nucleic Acids Res., 41, 2013
|
|
3IXN
 
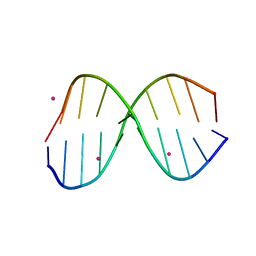 | |
3WTD
 
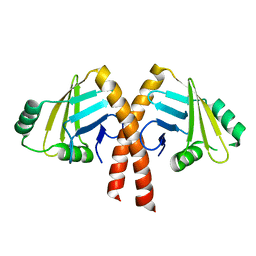 | | Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2014-04-09 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair.
Science, 347, 2015
|
|
1Z3F
 
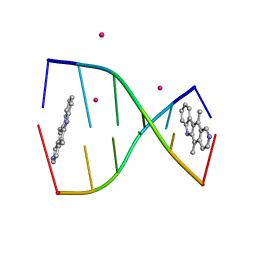 | | Structure of ellipticine in complex with a 6-bp DNA | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*G)-3', COBALT (II) ION, ELLIPTICINE | | Authors: | Canals, A, Purciolas, M, Aymami, J, Coll, M. | | Deposit date: | 2005-03-12 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The anticancer agent ellipticine unwinds DNA by intercalative binding in an orientation parallel to base pairs.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1QXB
 
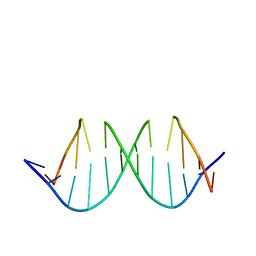 | | NMR structure determination of the self complementary DNA Dodecamer CGCGAATT*CGCG in which a ribose is inserted between the 3'-OH of T8 and the 5'-phosphate group of C9 | | Descriptor: | 5'-d(CpGpCpGpApApTpTpCpGpCpG)-3', beta-D-ribofuranose | | Authors: | Nauwelaerts, K, Vastmans, K, Froeyen, M, Kempeneers, V, Rozenski, J, Rosemeyer, H, Van Aerschot, A, Busson, R, Efimtseva, E, Mikhailov, S, Lescrinier, E, Herdewijn, P. | | Deposit date: | 2003-09-05 | | Release date: | 2004-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cleavage of DNA without loss of genetic information by incorporation of a disaccharide nucleoside.
Nucleic Acids Res., 31, 2003
|
|
3EY1
 
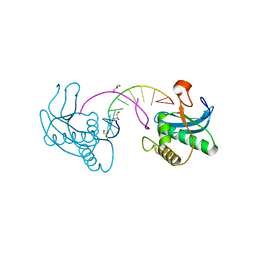 | |
3FQB
 
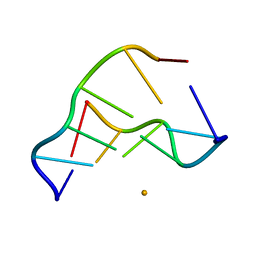 | |
1OSR
 
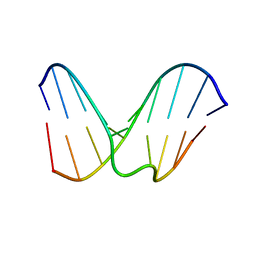 | | Structural study of dna duplex containaing a n-(2-deoxy-beta-erytho-pentofuranosyl) formamide frameshift by nmr and restrained molecular dynamics | | Descriptor: | 5'-D(*AP*GP*GP*AP*CP*CP*AP*CP*G)-3', 5'-D(*CP*GP*TP*GP*GP*(2DF)P*TP*CP*CP*T)-3' | | Authors: | Maufrais, C, Fazakerley, G.V, Cadet, J, Boulard, Y. | | Deposit date: | 2003-03-20 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural study of DNA duplex containing an N-(2-deoxy-beta-D-erythro-pentofuranosyl) formamide
frameshift by NMR and restrained molecular dynamics.
Nucleic Acids Res., 31, 2003
|
|
5YZE
 
 | | Crystal structure of the [Co2+-(chromomycin A3)2]-d(CCG)3 complex | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Chen, Y.W, Wu, P.C, Satange, R.B. | | Deposit date: | 2017-12-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | CoII(Chromomycin)2 Complex Induces a Conformational Change of CCG Repeats from i-Motif to Base-Extruded DNA Duplex
Int J Mol Sci, 19, 2018
|
|
2W36
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*BRUP*AP*CP*IP*GP*AP*BRUP*CP*GP)-3', ENDONUCLEASE V | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2W35
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*AP*GP*CP*CP*GP*TP)-3', 5'-D(*AP*TP*GP*CP*GP*AP*CP*IP*GP)-3', Endonuclease V, ... | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
