8E1H
 
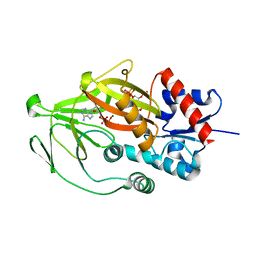 | | Asp1 kinase in complex with ADP Mg 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8JZN
 
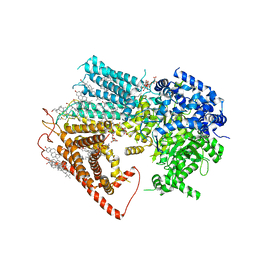 | | Structure of a fungal 1,3-beta-glucan synthase | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, C, You, Z, Chen, D, Hang, J, Wang, Z, Meng, J, Wang, L, Zhao, P, Qiao, J, Yun, C, Bai, L. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structure of a fungal 1,3-beta-glucan synthase.
Sci Adv, 9, 2023
|
|
8E1I
 
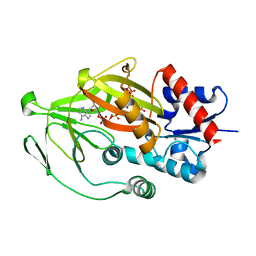 | | Asp1 kinase in complex with ATP Mg 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-TRIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1S
 
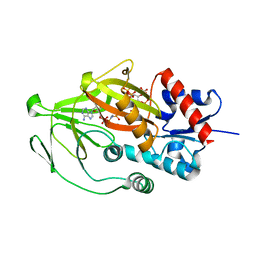 | | Asp1 kinase in complex with ADPNP Mn IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MANGANESE (II) ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1T
 
 | | Asp1 kinase in complex with ADPNP Mg IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MAGNESIUM ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1J
 
 | | Asp1 kinase in complex with 1,5-IP8 | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E1V
 
 | | Asp1 kinase in complex with ADPNP Mg IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, MAGNESIUM ION, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8HMB
 
 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with benidipine (BEN) | | Descriptor: | (3R)-1-benzylpiperidin-3-yl methyl (2R,3R,4R,5R,6S)-2,6-dimethyl-4-(3-nitrophenyl)piperidine-3,5-dicarboxylate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HMA
 
 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with tetrandrine (TET) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6,6',7,12-tetramethoxy-2,2'-dimethyl-1beta-3,4-didehydroberbaman, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HFF
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of norepinephrine in an inward-open state at resolution of 2.9 angstrom. | | Descriptor: | L-NOREPINEPHRINE, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
8HFL
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant bupropion in an inward-open state at resolution of 3.0 angstrom. | | Descriptor: | Bupropion, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
8HFI
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant desipramine in an inward-open state at resolution of 2.5 angstrom. | | Descriptor: | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
8HFG
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of dopamine in an inward-open state at resolution of 3.0 angstrom. | | Descriptor: | CHLORIDE ION, L-DOPAMINE, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
8HFE
 
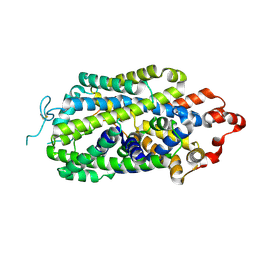 | | Cryo-EM structure of human norepinephrine transporter NET in an inward-open state at resolution of 2.5 angstrom | | Descriptor: | CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
7URO
 
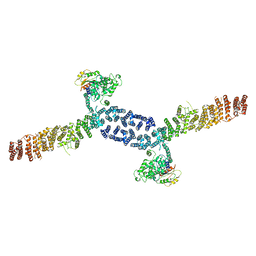 | |
7URR
 
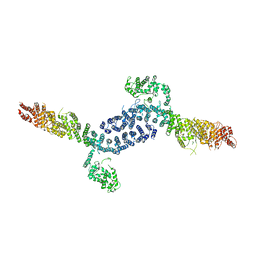 | |
8E20
 
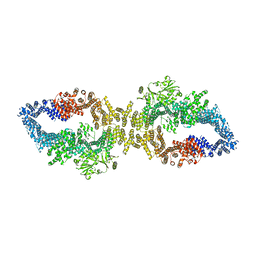 | |
7UX3
 
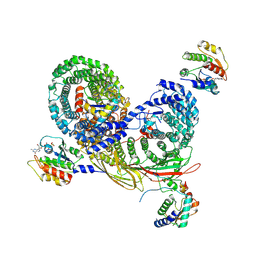 | | Asymmetric unit of AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated narrow membrane tubes | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-05-04 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8ECR
 
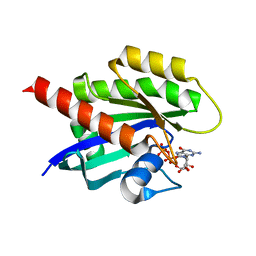 | | KRAS4b WT 1-185 bound to GDP-Mg2+ | | Descriptor: | GLYCEROL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Betts, L, Rossman, K.L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | KRAS4b WT 1-185 bound to GDP-Mg2+
To Be Published
|
|
8EDY
 
 | |
8EDN
 
 | |
8EDL
 
 | |
8EER
 
 | |
8EDM
 
 | |
8EHG
 
 | | Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
